Embed presentation
Download to read offline
![BABA SAHEB BHIMRAO
AMBEDKAR UNIVERSITY
PRESENTATION ON
ALGORITHM
BY :- PRASHANT TRIPATHI
M.Sc[BBAU]](https://image.slidesharecdn.com/vmsirpptpart1-170207032507/85/PPT-ON-ALGORITHM-1-320.jpg)

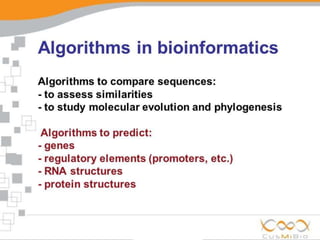
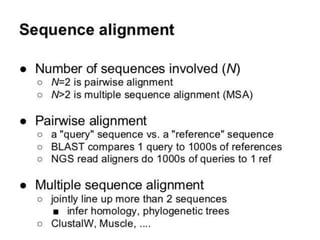

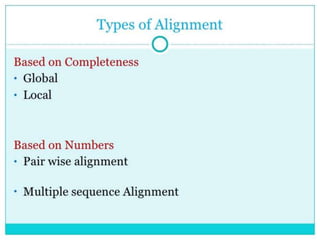

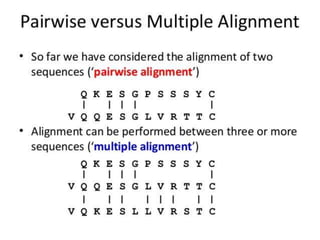

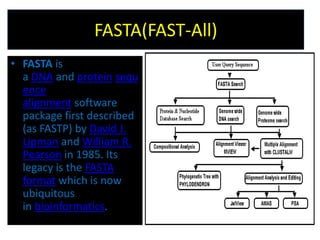

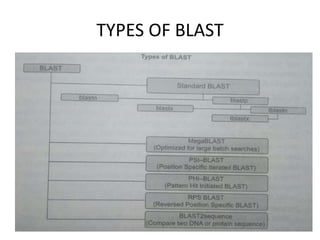
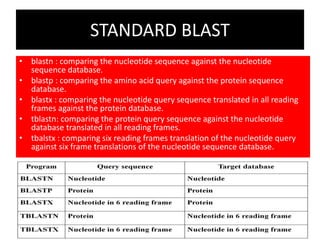

An algorithm is a finite set of steps to solve a problem or achieve a goal. It takes initial input and proceeds through a series of well-defined states until it reaches an output and terminates. FASTA is a widely used DNA and protein sequence alignment software that was first described in 1985. It uses a ubiquitous format for bioinformatics. There are different types of BLAST algorithms for comparing nucleotide and protein sequences.
![BABA SAHEB BHIMRAO
AMBEDKAR UNIVERSITY
PRESENTATION ON
ALGORITHM
BY :- PRASHANT TRIPATHI
M.Sc[BBAU]](https://image.slidesharecdn.com/vmsirpptpart1-170207032507/85/PPT-ON-ALGORITHM-1-320.jpg)












