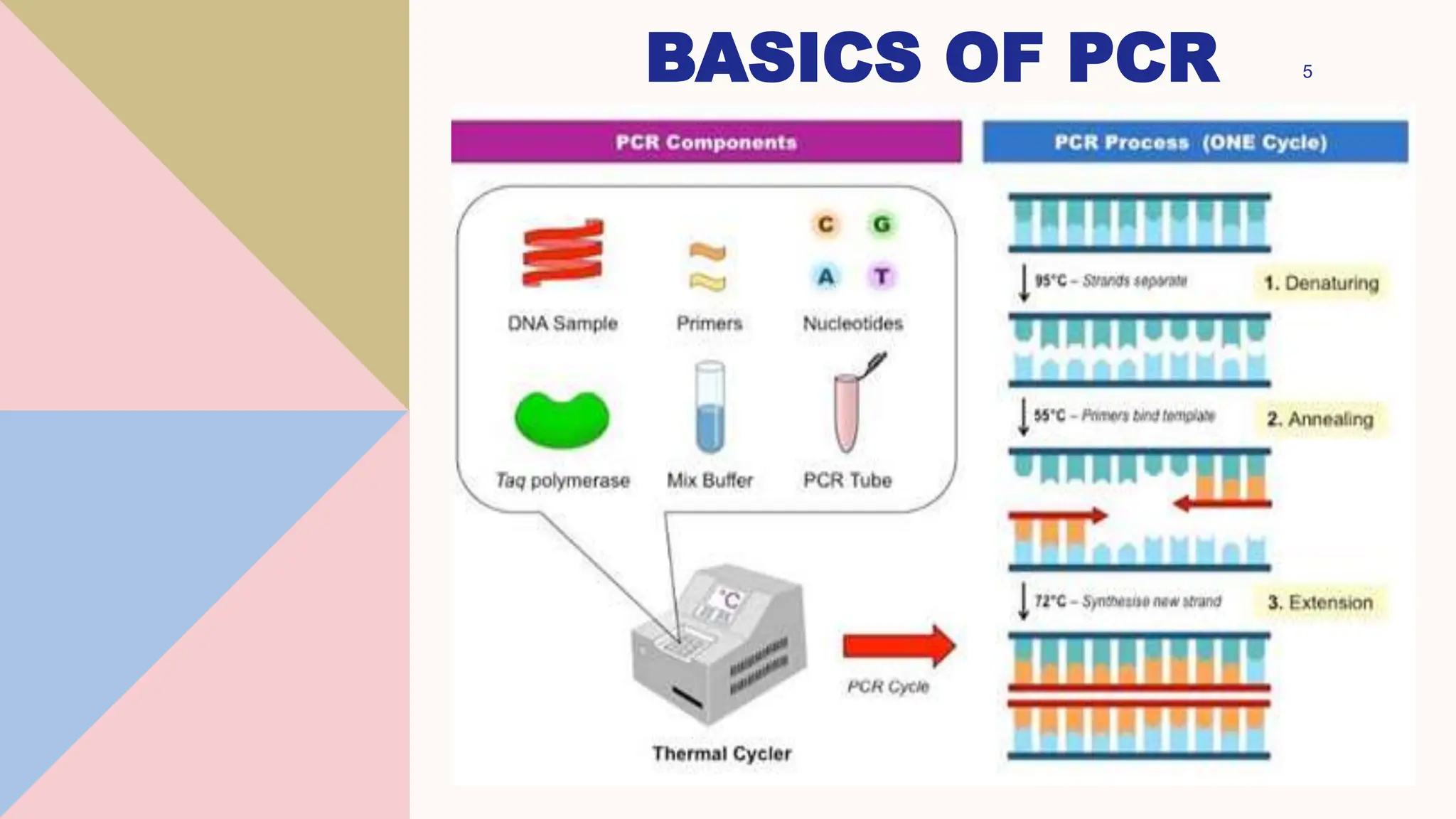
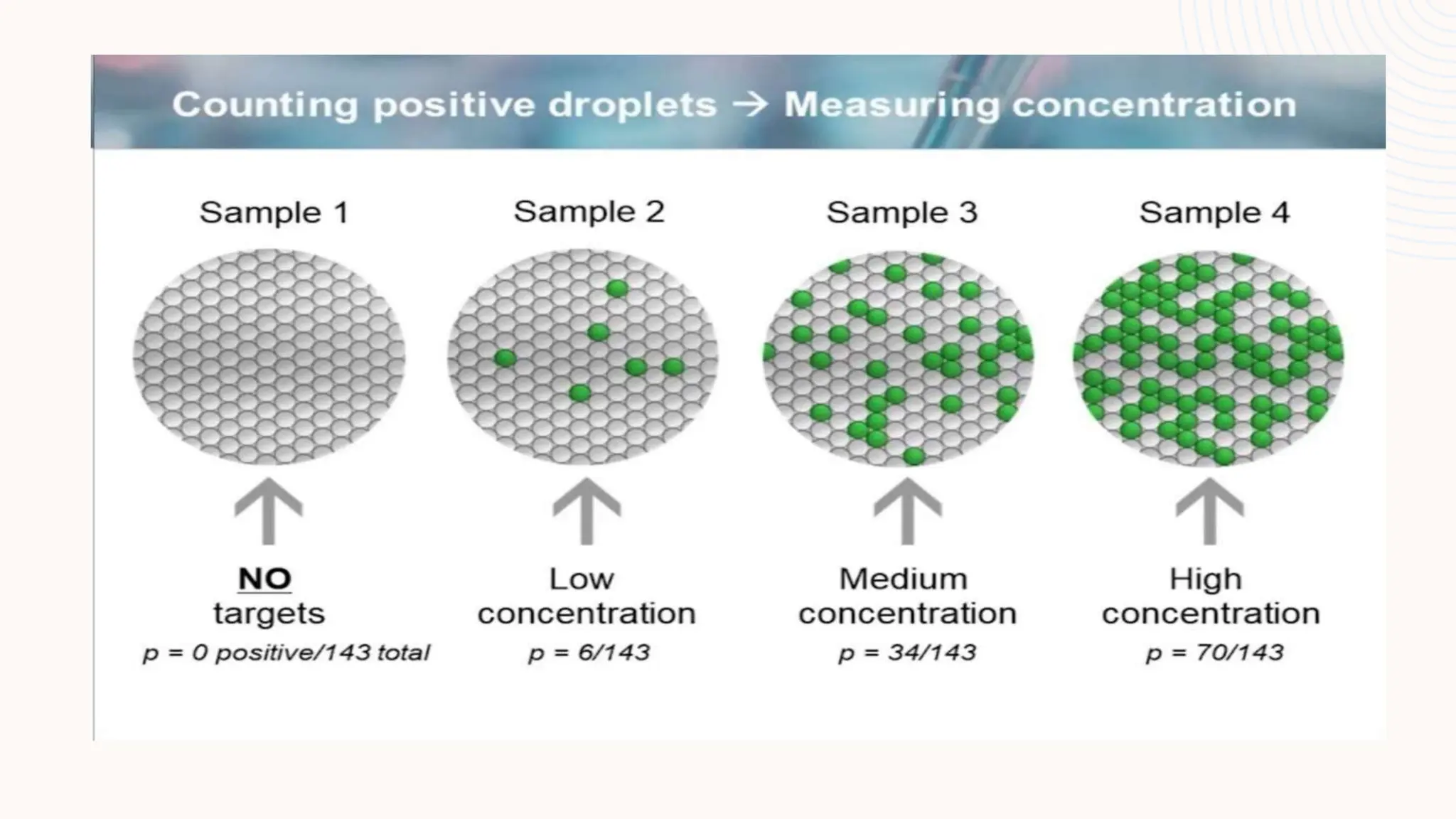
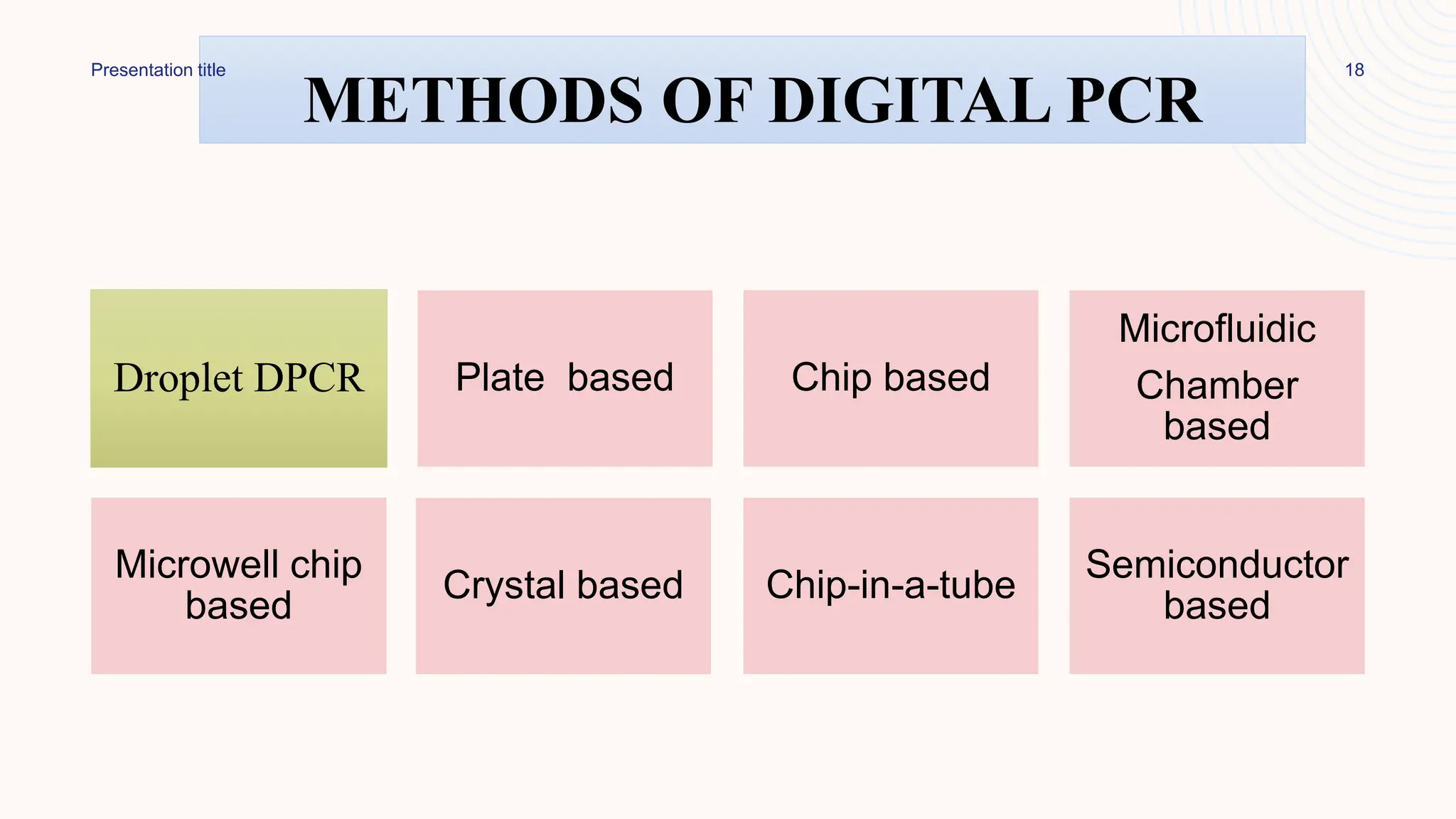
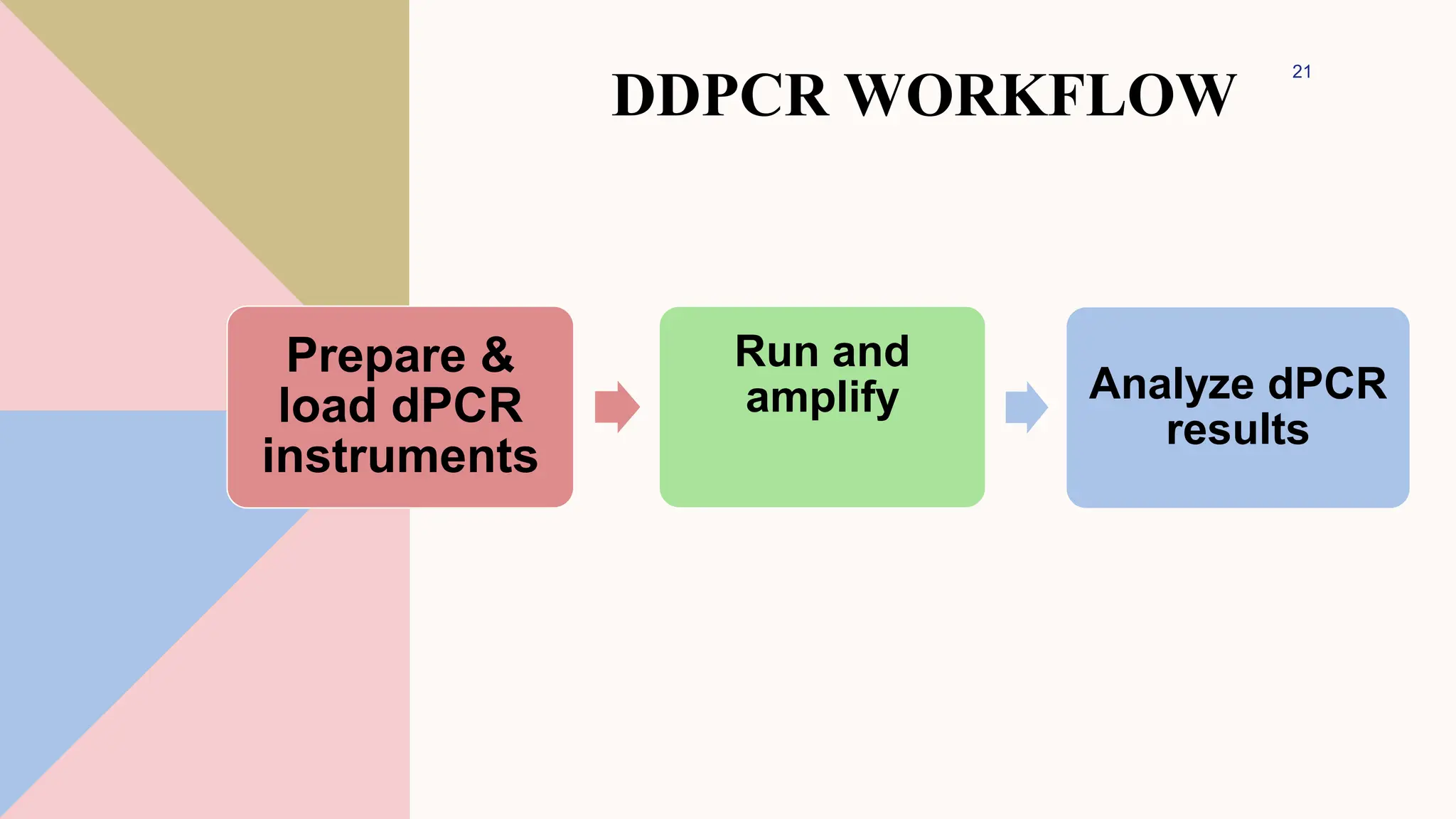
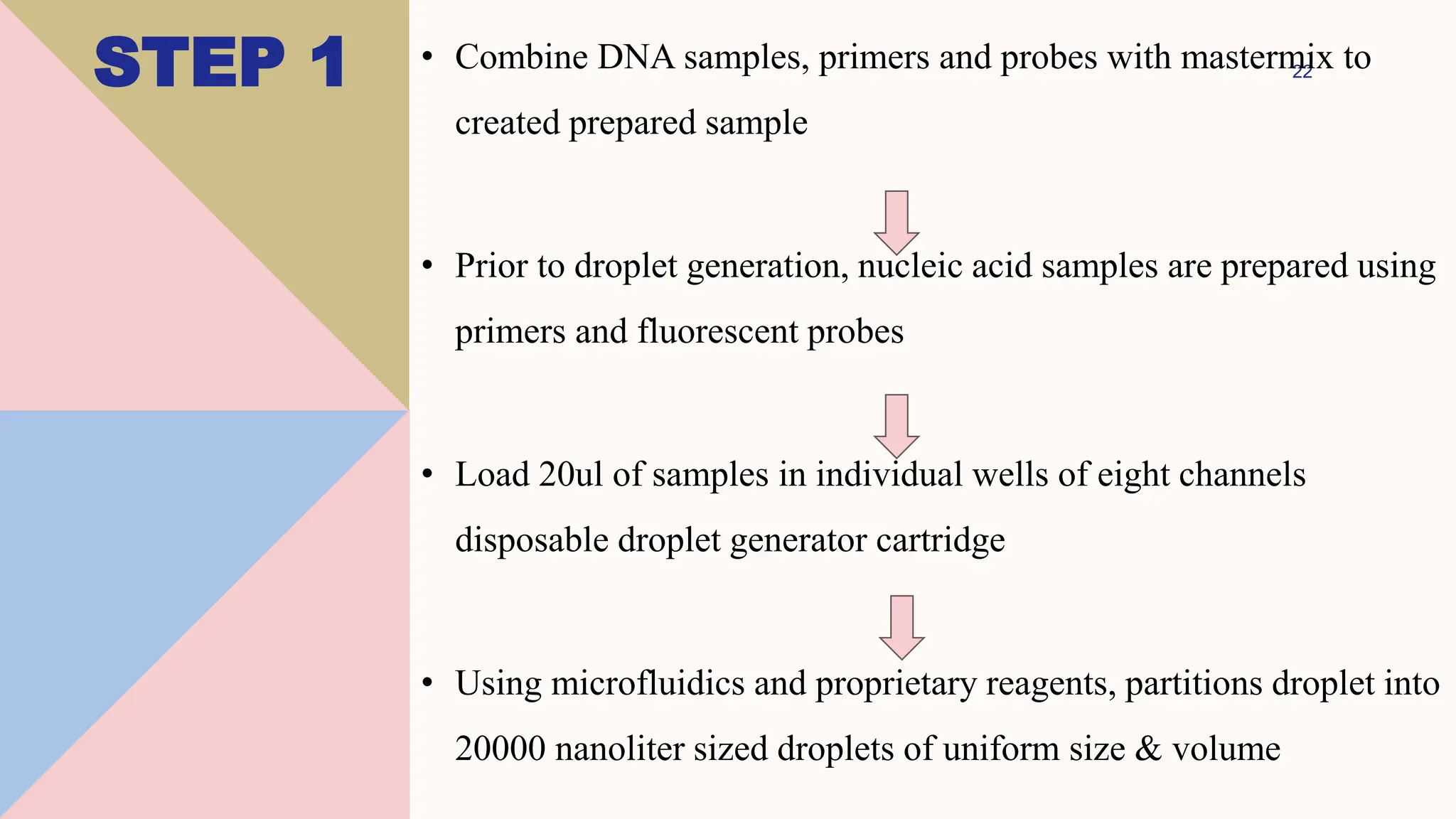
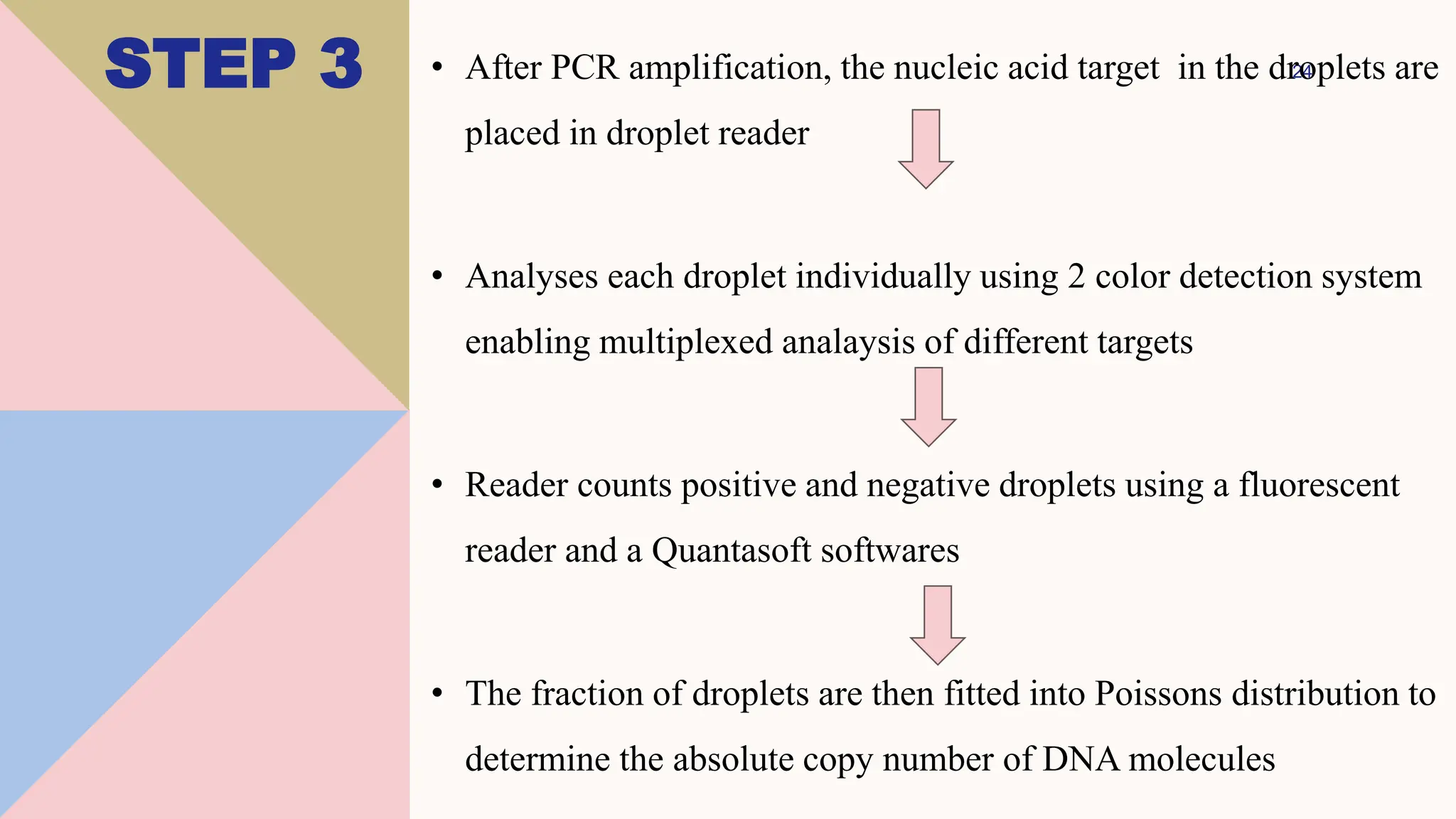
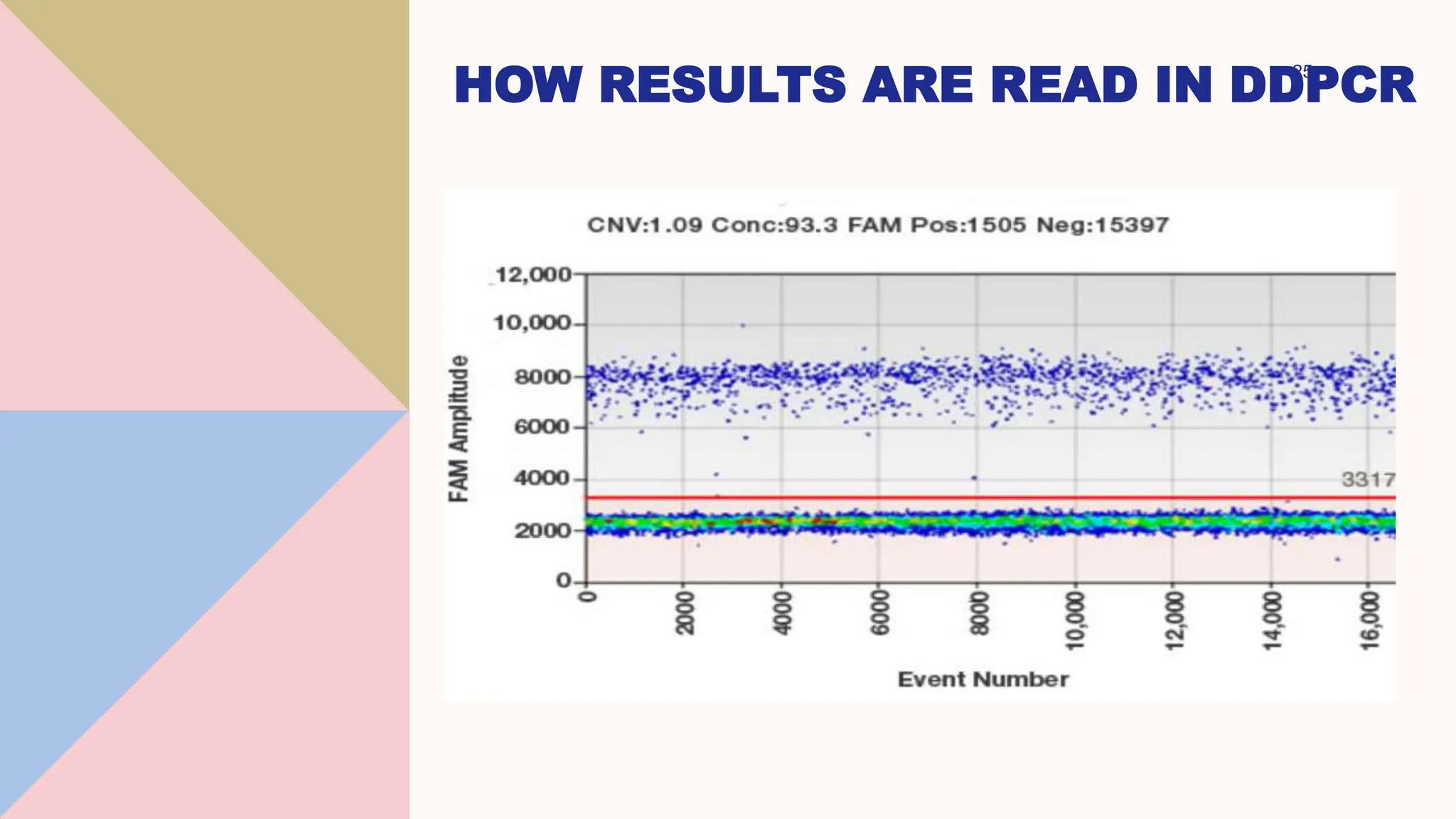
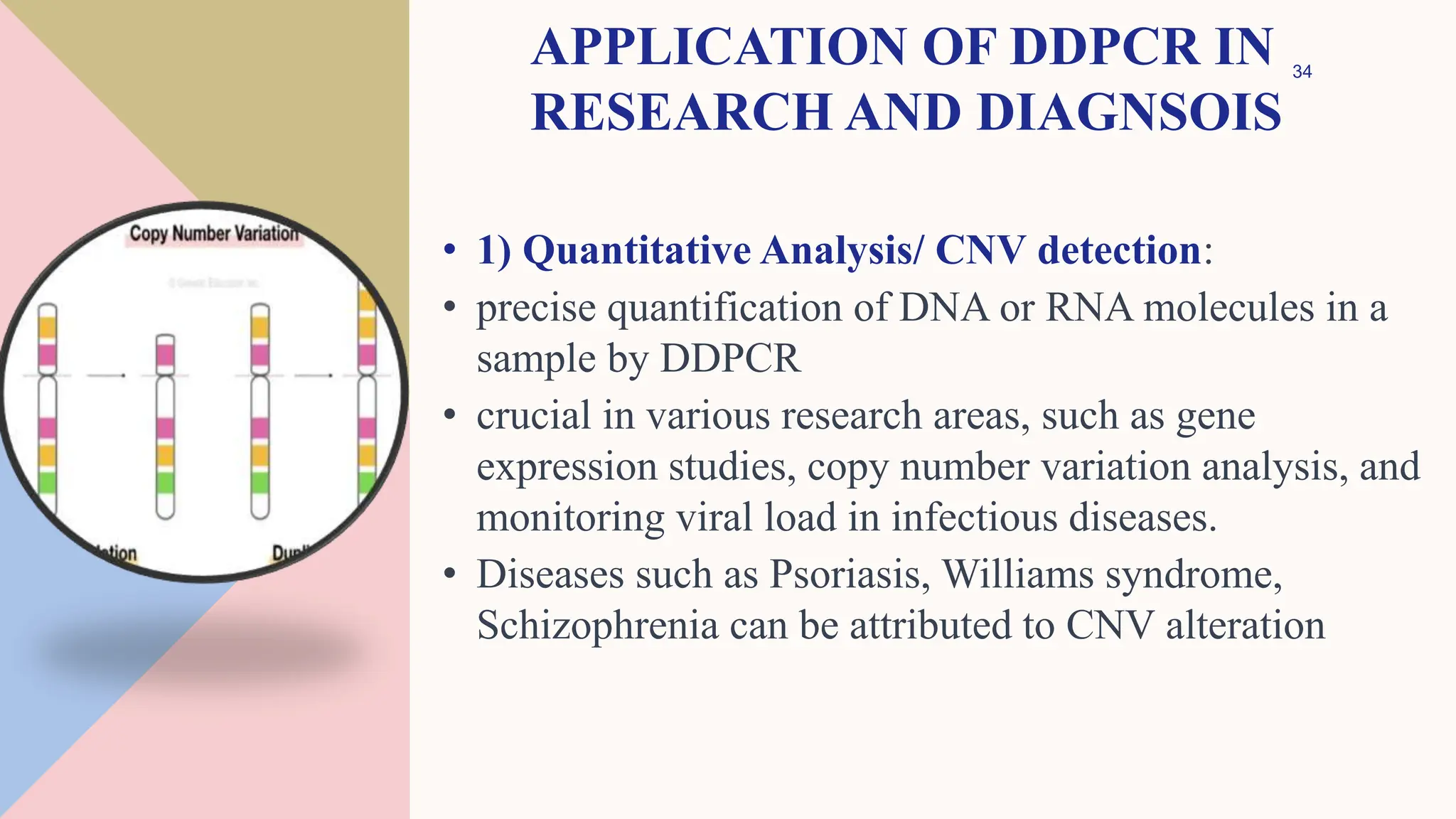
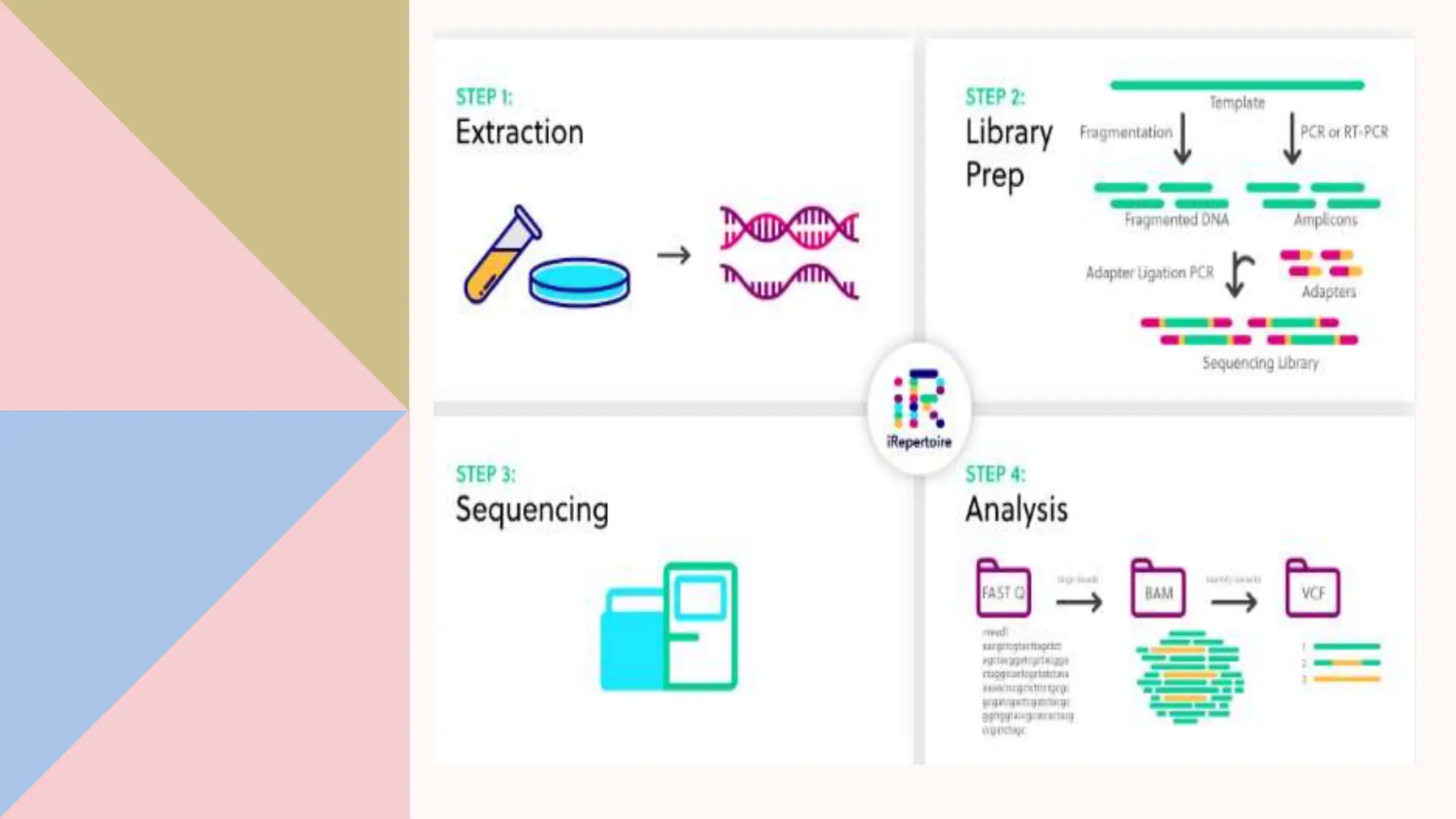
Digital droplet PCR (DDPCR) is a technique that partitions a sample into thousands of oil-encapsulated droplets for PCR amplification to provide an absolute quantification of target nucleic acids. DDPCR has applications in research and diagnosis such as precise quantification, rare mutation detection in liquid biopsies, genome editing detection, NGS library quantification, and analysis of gene and miRNA expression. Key benefits of DDPCR include improved sensitivity and multiplexing, high reproducibility, and providing absolute rather than relative quantification.