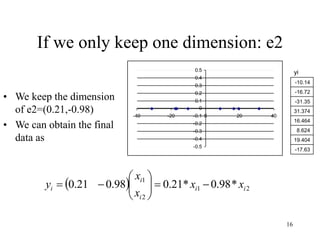
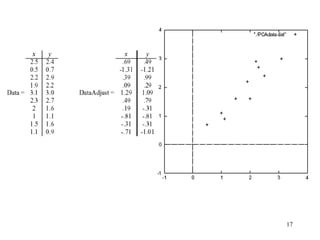
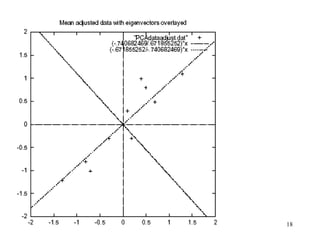
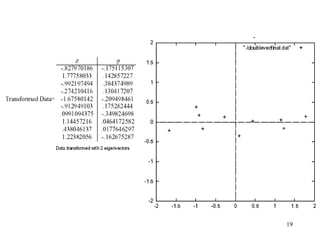
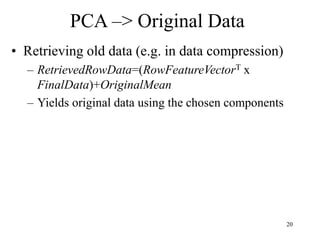
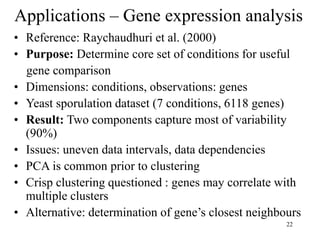
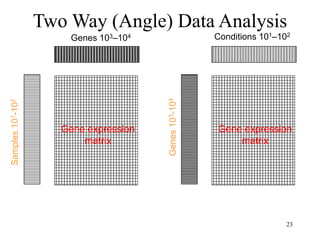
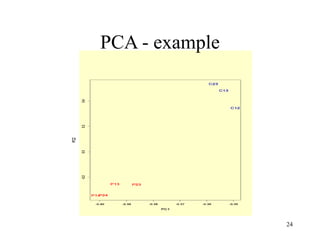
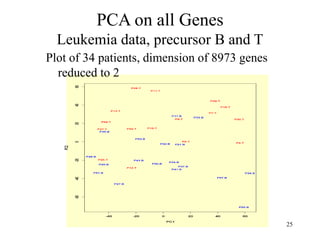
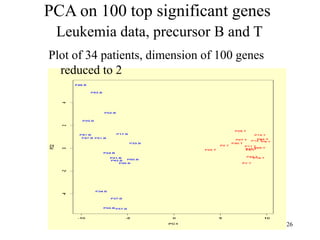
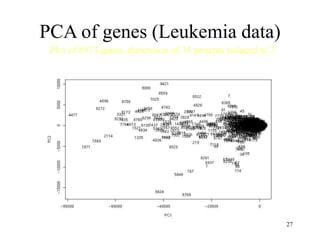
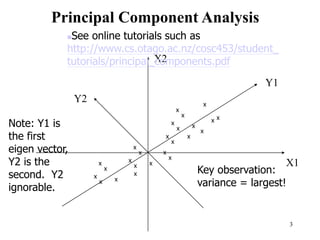
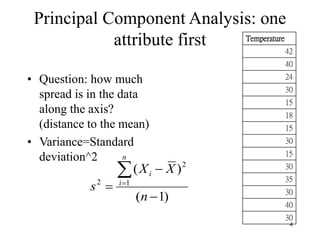
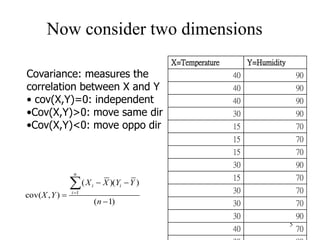
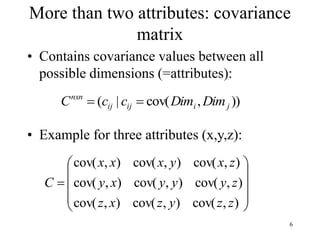
Principal Components Analysis (PCA) is an exploratory technique used to reduce the dimensionality of data sets while retaining as much information as possible. It transforms a number of correlated variables into a smaller number of uncorrelated variables called principal components. PCA is commonly used for applications like face recognition, image compression, and gene expression analysis by reducing the dimensions of large data sets and finding patterns in the data.
![11
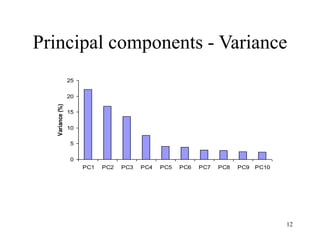
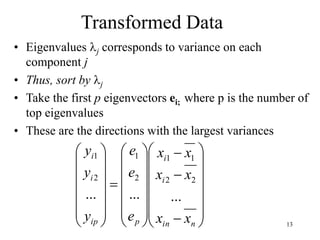
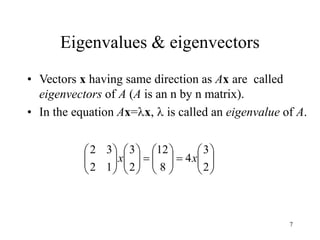
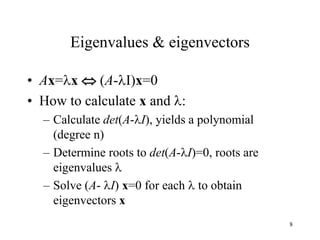
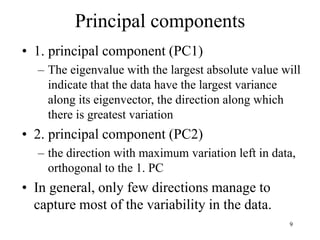
Eigenvalues
• Calculate eigenvalues and eigenvectors x for
covariance matrix:
– Eigenvalues j are used for calculation of [% of total
variance] (Vj) for each component j:
n
x
x
n
x
x
j
j n
V
1
1
100
](https://image.slidesharecdn.com/pca-230220075623-bfcf9c29/85/pca-ppt-11-320.jpg)