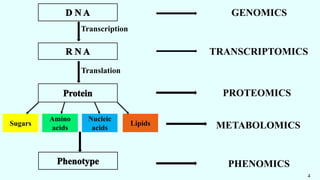
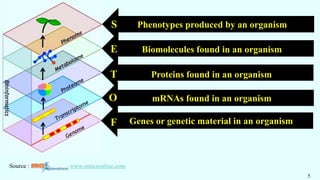
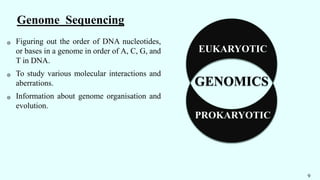
The document discusses the role of various 'omics' technologies—genomics, transcriptomics, proteomics, metabolomics, and phenomics—in crop improvement, emphasizing comprehensive analysis for developing improved organisms. It covers methodologies for mapping genes and traits, highlights achievements in nutrient enrichment of crops, and examines applications and limitations of these technologies. The information presents a forward-looking view on the integration of genomic data and next-generation sequencing for enhancing agricultural productivity and addressing biotic and abiotic stresses.


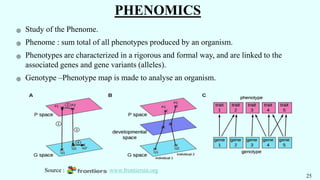
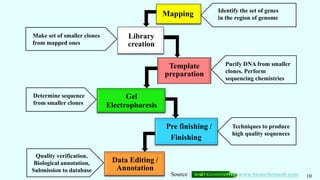
![Genome Mapping
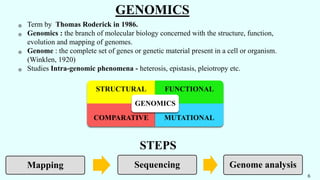
֍ Methods used to identify the locus of a gene and the distances between genes.
֍ Given by Alfred Sturtvent (1915) in Drosophila melanogaster.
֍ Traits mapped : Morphological Characters, Productivity traits, Resistance Traits,
Quality Traits, Agronomic Traits, Special Characters.
֍ To study linkage and recombination.
֍ In India, by the Depart of Biotechnology [DBT] and ICAR.
֍ Done in rice, wheat, maize, chick pea, banana, tomato, Brassica, etc.
7
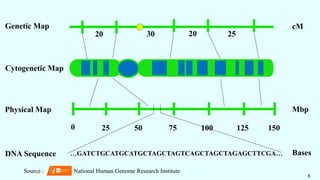
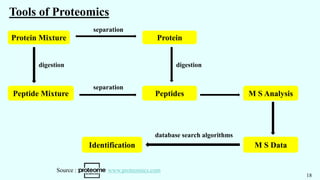
MAPPING
GENETIC
MAP
CYTOGENETIC
MAP
PHYSICAL
MAP](https://image.slidesharecdn.com/omics-230515052443-672899c5/85/OMICS-in-Crop-Improvement-pptx-7-320.jpg)






![1. Metabolite Extraction 2. Chromatography 3. Mass Spectrometry 4. Data Analysis
[ Rapid Enzyme Quenching ] [ Separation ] [ Specificity ] [ Metabolite identification ]
Cell & Tissues
Leaves & other organs
Cold organic
solvents
Grinding
Weighing
Samples
23
Source : www.metabolon.com](https://image.slidesharecdn.com/omics-230515052443-672899c5/85/OMICS-in-Crop-Improvement-pptx-23-320.jpg)