New methods for high-throughput nucleic sequencing and diagnostics using a thermostable group II intron reverse transcriptase (TGIRT)
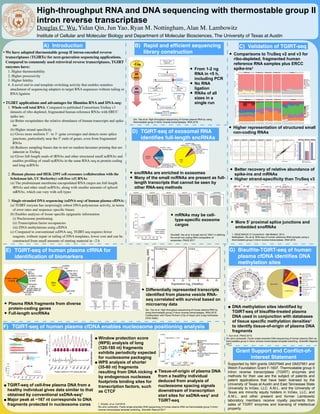
- 1. ß High-throughput RNA and DNA sequencing with thermostable group II intron reverse transcriptase Douglas C. Wu, Yidan Qin, Jun Yao, Ryan M. Nottingham, Alan M. Lambowitz Institute of Cellular and Molecular Biology and Department of Molecular Biosciences, The University of Texas at Austin • We have adapted thermostable group II intron-encoded reverse transcriptases (TGIRTs) for next-generation sequencing applications. Compared to commonly used retroviral reverse transcriptases, TGIRT enzymes have: 1. Higher thermostability 2. Higher processivity 3. Higher fidelity 4. A novel end-to-end template-switching activity that enables seamless attachment of sequencing adapters to target RNA sequences without tailing or RNA ligation • TGIRT applications and advantages for Illumina RNA and DNA-seq: 1. Whole-cell total RNA. Compared to published Consortium TruSeq v3 datasets of ribo-depleted, fragmented human reference RNAs with ERCC spike-ins: (a) Better recapitulates the relative abundance of human transcripts and spike- ins (b) Higher strand-specificity (c) Gives more uniform 5’- to 3’-gene coverages and detects more splice junctions, particularly near the 5’ ends of genes, even from fragmented RNAs (d) Reduces sampling biases due to not-so-random hexamer priming that are inherent in TruSeq (e) Gives full-length reads of tRNAs and other structured small ncRNAs and enables profiling of small ncRNAs in the same RNA-seq as protein-coding and long ncRNAs 2. Human plasma and HEK-239T cell exosomes (collaboration with the Schekman lab, UC Berkerley) cell-free (cf) RNAs: (a) The predominant membrane-encapsulated RNA cargos are full-length tRNAs and other small ncRNAs, along with smaller amounts of spliced mRNAs, which can vary with cell types 3. Single-stranded DNA sequencing (ssDNA-seq) of human plasma cfDNA: (a) TGIRT enzyme has surprisingly robust DNA polymerase activity, in terms of error rates and sequence-specific biases (b) Enables analysis of tissue-specific epigenetic information (i) Nucleosome positioning (ii) Transcription factor occupancies (iii) DNA methylations using cfDNA (c) Compared to conventional ssDNA-seq, TGIRT-seq requires fewer reagents, without repair or tailing of DNA templates, lower cost and can be constructed from small amounts of starting material in ~2 h B) Rapid and efficient sequencing library construction • Higher representation of structured small non-coding RNAs • Better recovery of relative abundance of spike-ins and mRNAs • Higher strand-specificity than TruSeq v3 167 nt 0.0 0.5 1.0 1.5 2.0 2.5 0 50 100 150 200 250 300 350 400 Fragment length (nt) TGIRT−seq ssDNA−seq Long(120−180nt)Short(35−80nt) −1000 −800 −600 −400 −200 0 200 400 600 800 1000 −2 −1 0 1 2 −4 0 4 8 Position relative to CTCF binding sites (bp) ScaledWPSs TGIRT−seq ssDNA−seq −120 −100 −80 −60 −40 −20 0 20 40 60 80 100 −120 −100 −80 −60 −40 −20 0 20 40 60 80 100 Position relative to center of 167−nt fragments (bp) AA/AT/TA/TT GG/GC/CG/CC b N ~340 bp ~167 bp ~145 bp Core histone Peripheral histone H1 Linker DNA 3’ blocker 5’ R2 RNA R2R DNA TGIRT 3’ 5’3’ 5’ 3’ 5’ 3’ 5’ 3’ 5’ 3’ 5’ 3’ 5’ 3’ 5’ 167 nt 0.0 0.5 1.0 1.5 2.0 2.5 0 50 100 150 200 250 300 350 400 Fragment length (nt) %Reads TGIRT−seq ssDNA−seq 10.4 nt 0 1 2 3 0 100 200 300 400 Inter−nucleosome distance in cfDNA of two individuals (bp) %Peaks TGIRT−seq ssDNA−seq 180 bp 0 2 4 −720−600−480−360−240−120 0 120 240 360 480 600 720 Inter−nucleosome distance between different male individual analyzed by TGIRT−seq and ssDNA−seq (bp) Peakcount(x106 ) Testis HEL Spearman's ρ = 0.83 0.000 0.005 0.010 0.015 0.00 0.01 0.02 0.03 0.04 Pearson's ρ (ssDNA−seq) Pearson'sρ(TGIRT−seq) Abdominal Brain Breast/Female Reproductive Lung Lymphoid Myeloid Sarcoma Skin Urinary/Male Reproductive Primary Tissue Other a b c Figure 4 •Window protection score (WPS) analysis of long (120-180 nt) fragments exhibits periodicity expected for nucleosome packaging •WPS analysis of shorter (35-80 nt) fragments resulting from DNA nicking by endogenous nucleases footprints binding sites for transcription factors, such as CTCF •TGIRT-seq of cell-free plasma DNA from a healthy individual gives data similar to that obtained by conventional ssDNA-seq1 •Major peak at ~167 nt corresponds to DNA fragments protected in nucleosome cores Supported by NIH grants GM37949 and GM37951 and Welch Foundation Grant F-1607. Thermostable group II intron reverse transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas at Austin and East Tennessee State University to InGex, LLC. A.M.L. and the University of Texas are minority equity holders in InGex, LLC, and A.M.L. and other present and former Lambowitz laboratory members receive royalty payments from sales of TGIRT enzymes and licensing of intellectual property. Neutrophils: 50% Lymphocytes: 21% Lungs: 10% Adipose tissues: 9.4% Liver: 6.6% Heart: 1.7% Brain: 0.67% Small intestines: 4.8e−18% Adrenal glands: 0% Colon: 0% Esophagus: 0% Pancreas: 0% a b Neturophils Lymphocytes Lung Adipose tissues Liver 0M 50M 100M 150M 200M 0M 50M 100M150M 200M 0M 50M 100M 150M 0M 50M 100M 150M 0M 50M 100M 150M 0M 50M 100M150M 0M 50M 100M 150M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 0M 50M 0M 50M 0M 50M 0M 50M 0M 0M 50M chr1 chr2 chr3 chr4chr5 chr6 chr7 chr8chr9 chr10 chr11 chr12 chr13 chr14chr15chr16 chr17 chr18 chr19 chr20chr21chr22 Neutrophils Lymphocytes Adipose tissues Pancreas Adrenal glands Lungs Heart Liver Colon Brain Small intestines Esophagus Neutrophils: 50% Lymphocytes: 21% Lungs: 10% Adipose tissues: 9.4% Liver: 6.6% Heart: 1.7% Brain: 0.67% Small intestines: 4.8e−18% Adrenal glands: 0% Colon: 0% Esophagus: 0% Pancreas: 0% a b Neturophils Lymphocytes Lung Adipose tissues Liver 0M 50M 100M 150M 200M 0M 50M 100M150M 200M 0M 50M 100M 150M 0M 50M 100M 150M 0M 50M 100M 150M 0M 50M 100M150M 0M 50M 100M 150M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 100M 0M 50M 0M 50M 0M 50M 0M 50M 0M 50M 0M 0M 50M chr1 chr2 chr3 chr4chr5 chr6 chr7 chr8chr9 chr10 chr11 chr12 chr13 chr14chr15chr16 chr17 chr18 chr19 chr20chr21chr22 Neutrophils Lymphocytes Adipose tissues Pancreas Adrenal glands Lungs Heart Liver Colon Brain Small intestines Esophagus • DNA methylation sites identified by TGIRT-seq of bisulfite-treated plasma DNA used in conjunction with databases of tissue specific methylation densities1 to identify tissue-of-origin of plasma DNA fragments Cell Exo Cell Exo HEK293T 100,000Xg Pellet Resuspend 60% sucrose 20% 40% 60% 150,000Xg 1,500Xg 10,000Xg Supernatant Wash Pellet 120,000Xg TGIRT-seq Media anti-CD63 beads A B D E C 100 101 102 103 104 105 106 100 101 102 103 104 105 106 WT / mRNAs Whole cell (Fragmented) Exosomes r = 0.64 5’ TOP mRNAs aaRS mRNAs Other mRNAs 7SK (88) 18/28S rRNA (2) 5/5.8S rRNA (2) Antisense (868) lincRNA (1,332) sncRNA (2,221) Mt (37) Other lncRNA (349) Protein coding (10,767) Pseudogene (1.358) Y RNA (689) Other sncRNA (4) 7SL (325) miRNA (263) snoRNA (285) snRNA (510) tRNA (53) Vault RNA (4) EVs Cells RNaseI RNaseI+TX-100100Kpellet MIF RPS2 RPS17 334 bp 489 bp 1,000 500 300 500 200 100 500 300 200 347 bp EEF1A1 EEF2 HSP90AA1 NPM1 RPS6 ENO1 RPS4X PRKDC NUCKS1 EEF1G RPL4 PARP1 HNRNPU HSP90AB1 HSPA1B TUBB TUBA1B DDX17 DYNC1H1 RPL5 CANX HIST1H1C RPSA SERBP1 HIST1H1E HSPD1 ACTB IRS4 SCD LDHB PRPF8 AMOT ACTG1 HSPA1A PLS3 HUWE1 EIF4G2 HNRNPA2B1 RPS18 HNRNPK YBX1 RPS2 SET NCL RPL3 RPS17 CLTC KPNB1 PTMA RPL13 4 8 12 EEF1A1 RPL5 H3F3A RPL39 RPS14 HSP90AA1 RPS6 NCL NPM1 SERBP1 B4GALNT2 HIST1H1E RPS2 RPL12 RPS17 RPL11 ACTB RPL7 RPL6 YBX1 EEF2 ANP32B NET1 RPL14 RPL23 RAB13 RPS3A ATP5B ENO1 HIST2H4B RPL27 RPL37A MTRNR2L12 RPS3 HIST3H2A RPLP1 RPS18 HIST2H2AA4 RPS19 RPS11 ATAD5 PARP1 PTMA RPSA HIST2H2AA3 HIST1H2AG HIST1H2BK RPS7 RPS4X EIF3A peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. The copyright holder for this preprint (which was not.http://dx.doi.org/10.1101/160556doi:bioRxiv preprint first posted online Jul. 7, 2017; • From 1-2 ng RNA in <5 h, including PCR • No RNA ligation • RNAs of all sizes in a single run C) Validation of TGIRT-seq D) TGIRT-seq of exosomal RNA identifies full-length sncRNAs E) TGIRT-seq of human plasma cfRNA for identification of biomarkers F) TGIRT-seq of human plasma cfDNA enables nucleosome positioning analysis G) Bisulfite-TGIRT-seq of human plasma cfDNA identifies DNA methylation sites A) Introduction Shurtleff, Yao et al. A broad role for YBX1 in defining the small non-coding RNA composition of exosomes. PNAS 2017 Qin, Yao et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016 1. SEQC/MAQC-III Consortium. Nat Biotech. 2014 Nottingham, Wu et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 2016 Qin, Yao et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016 Collaboration with Flavia Pichiorri (City of Hope) and Craig Hofmeister (Ohio State) Grant Support and Conflict-of- interest Statement 1. Sun et al. PNAS 2015 Wu and Lambowitz. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Scientific Reports 2017 the three methods was similar with TGIRT-seq having a roughly twofold lower limit of detection when compared to the TruSeq libraries at a threshold of 1 FPKM (fragments per kilobase per million mapped reads). TruSeq v2 libraries had a slightly higher number of detected spike-in species, likely due to their greater sequencing depth (Supplemental Table S1). Second, each of the 92 polyadenylated ERCC spike-in tran- scripts is grouped into one of four classes (0.5:1, 0.67:1, 1:1, 4:1) according to the relative abundance of the spike-in be- tweenMix 1(Sample A)and Mix 2(Sample B).TGIRT-seq re- capitulated these differences in abundance better than the strand-specific TruSeq v3 method and almost as well as the non-strand-specific TruSeq v2 method (Fig. 3B). For TGIRT-seq and TruSeq v2, empirical fold-change ratios were more highly correlated with their expected values for abundant spike-ins (those to the right of each panel), as previ- ously observed for TruSeq v2 (SEQC/MAQC-III Consortium 2014), whereas empirical fold-change ratios were poorly cor- related with their expected values for TruSeq v3 (Fig. 3B). Third, the mixing of Samples A and B to constitute Samples C and D defines an expected order of dilution of the human reference set RNAs. For both TGIRT-seq and TruSeq v3 (Samples C and D were not analyzed by TruSeq v2 in the ABRF study), most protein-coding gene transcripts followed a consistent titration order, with those following in- consistent order corresponding to transcripts with small fold changes between Samples A and B (Fig. 4A). For both meth- ods, there was also a slight bias toward inconsistent titration order for transcripts higher in B than in A (tail on right side of the red peak). More detailed analysis of protein-coding gene transcripts detected by TGIRT-seq and TruSeq v3 in Samples A–D (Fig. 4B) revealed that both protocols performed similarly in recovering the known mixing ratios between samples. The TGIRT-seq libraries had an observed mixing ratio FIGURE 2. TGIRT-seq reads map mostly to protein-coding genes but with greater representation of small ncRNAs than TruSeq libraries. (A) Stacked bar graphs showing the percentage of uniquely mapped reads for each class of annotated genomic features in Ensembl GRCh38 release 76, Genomic tRNA Database, and piRNABank (Qin et al. 2016) for different library preparation methods for numbered replicates of Samples A–D. (B) Stacked bar graphs showing the percentage of small noncoding RNA reads that map to different classes of small ncRNAs for different library preparation methods for numbered replicates of Samples A–D. MiscRNA includes ribozymes, such as RNase P RNA, imprinted transcripts, such as Xist, and other tran- scripts that cannot be classified into other RNA annotation categories. (Left panels) TGIRT-seq; (middle panels) TruSeq v2 (from ABRF at three dif- ferent sites, L/R/V); (right panels) TruSeq v3 (from ABRF at site W). Features and small ncRNA classes are color coded as indicated to the right of the bar graphs. Nottingham et al. 6 RNA, Vol. 22, No. 4 Cold Spring Harbor Laboratory Presson January 29, 2016 - Published byrnajournal.cshlp.orgDownloaded from the three methods was similar with TGIRT-seq having a roughly twofold lower limit of detection when compared to the TruSeq libraries at a threshold of 1 FPKM (fragments per kilobase per million mapped reads). TruSeq v2 libraries had a slightly higher number of detected spike-in species, likely due to their greater sequencing depth (Supplemental Table S1). Second, each of the 92 polyadenylated ERCC spike-in tran- scripts is grouped into one of four classes (0.5:1, 0.67:1, 1:1, 4:1) according to the relative abundance of the spike-in be- tweenMix 1(Sample A)and Mix 2(Sample B).TGIRT-seq re- capitulated these differences in abundance better than the strand-specific TruSeq v3 method and almost as well as the non-strand-specific TruSeq v2 method (Fig. 3B). For TGIRT-seq and TruSeq v2, empirical fold-change ratios were more highly correlated with their expected values for abundant spike-ins (those to the right of each panel), as previ- ously observed for TruSeq v2 (SEQC/MAQC-III Consortium 2014), whereas empirical fold-change ratios were poorly cor- related with their expected values for TruSeq v3 (Fig. 3B). Third, the mixing of Samples A and B to constitute Samples C and D defines an expected order of dilution of the human reference set RNAs. For both TGIRT-seq and TruSeq v3 (Samples C and D were not analyzed by TruSeq v2 in the ABRF study), most protein-coding gene transcripts followed a consistent titration order, with those following in- consistent order corresponding to transcripts with small fold changes between Samples A and B (Fig. 4A). For both meth- ods, there was also a slight bias toward inconsistent titration order for transcripts higher in B than in A (tail on right side of the red peak). More detailed analysis of protein-coding gene transcripts detected by TGIRT-seq and TruSeq v3 in Samples A–D (Fig. 4B) revealed that both protocols performed similarly in recovering the known mixing ratios between samples. The TGIRT-seq libraries had an observed mixing ratio FIGURE 2. TGIRT-seq reads map mostly to protein-coding genes but with greater representation of small ncRNAs than TruSeq libraries. (A) Stacked bar graphs showing the percentage of uniquely mapped reads for each class of annotated genomic features in Ensembl GRCh38 release 76, Genomic tRNA Database, and piRNABank (Qin et al. 2016) for different library preparation methods for numbered replicates of Samples A–D. (B) Stacked bar graphs showing the percentage of small noncoding RNA reads that map to different classes of small ncRNAs for different library preparation methods for numbered replicates of Samples A–D. MiscRNA includes ribozymes, such as RNase P RNA, imprinted transcripts, such as Xist, and other tran- scripts that cannot be classified into other RNA annotation categories. (Left panels) TGIRT-seq; (middle panels) TruSeq v2 (from ABRF at three dif- ferent sites, L/R/V); (right panels) TruSeq v3 (from ABRF at site W). Features and small ncRNA classes are color coded as indicated to the right of the bar graphs. Nottingham et al. 6 RNA, Vol. 22, No. 4 Cold Spring Harbor Laboratory Presson January 29, 2016 - Published byrnajournal.cshlp.orgDownloaded from Cell Exo Cell Exo HEK293T 100,000Xg Pellet Resuspend 60% sucrose 20% 40% 60% 150,000Xg 1,500Xg 10,000Xg Supernatant Wash Pellet 120,000Xg TGIRT-seq Media anti-CD63 beads A B D E C 100 101 102 103 104 105 106 100 101 102 103 104 105 106 WT / mRNAs Whole cell (Fragmented) Exosomes r = 0.64 5’ TOP mRNAs aaRS mRNAs Other mRNAs 7SK (88) 18/28S rRNA (2) 5/5.8S rRNA (2) Antisense (868) lincRNA (1,332) sncRNA (2,221) Mt (37) Other lncRNA (349) Protein coding (10,767) Pseudogene (1.358) Y RNA (689) Other sncRNA (4) 7SL (325) miRNA (263) snoRNA (285) snRNA (510) tRNA (53) Vault RNA (4) EVs Cells RNaseI RNaseI+TX-100100Kpellet MIF RPS2 RPS17 334 bp 489 bp 1,000 500 300 500 200 100 500 300 200 347 bp EEF1A1 EEF2 HSP90AA1 NPM1 RPS6 ENO1 RPS4X PRKDC NUCKS1 EEF1G RPL4 PARP1 HNRNPU HSP90AB1 HSPA1B TUBB TUBA1B DDX17 DYNC1H1 RPL5 CANX HIST1H1C RPSA SERBP1 HIST1H1E HSPD1 ACTB IRS4 SCD LDHB PRPF8 AMOT ACTG1 HSPA1A PLS3 HUWE1 EIF4G2 HNRNPA2B1 RPS18 HNRNPK YBX1 RPS2 SET NCL RPL3 RPS17 CLTC KPNB1 PTMA RPL13 4 8 12 EEF1A1 RPL5 H3F3A RPL39 RPS14 HSP90AA1 RPS6 NCL NPM1 SERBP1 B4GALNT2 HIST1H1E RPS2 RPL12 RPS17 RPL11 ACTB RPL7 RPL6 YBX1 EEF2 ANP32B NET1 RPL14 RPL23 RAB13 RPS3A ATP5B ENO1 HIST2H4B RPL27 RPL37A MTRNR2L12 RPS3 HIST3H2A RPLP1 RPS18 HIST2H2AA4 RPS19 RPS11 ATAD5 PARP1 PTMA RPSA HIST2H2AA3 HIST1H2AG HIST1H2BK RPS7 RPS4X EIF3A peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. The copyright holder for this preprint (which was not.http://dx.doi.org/10.1101/160556doi:bioRxiv preprint first posted online Jul. 7, 2017; • sncRNAs are enriched in exosomes • Many of the small ncRNAs are present as full- length transcripts that cannot be seen by other RNA-seq methods Protein coding (19,683) Pseudogene (6,555) 18/28S rRNA (2) 5/5.8S rRNA (2) Antisense (4,862) lincRNA (6,580) sncRNA (1,725) Mt (37) Other lncRNA (1,387) tRNA (52) snRNA (114) snoRNA (190) miRNA (411) 7SL(295) 7SK(64) Other sncRNA (43) Y RNA (74) Vault RNA (4) miR-27a-3p LysCTT m1 A58 CCA Non-templated addition HisGTG m1 A585’ G m1 G37 ArgCCG m1 A58 m1 G37m1 A9 H/ACA box snoRNA SNORA9 Vault RNA 1-1 RN7SL2 5’ 5’ 5’ 5’ 5’ 5’3’ 3’ 3’ 3’3’ RNY4 5’ 3’ 3’ MIR16-2MIR142MIR451A MIR122 MIR27A miR-451a miR-27a-3p miR-142-3p miR-16-5p miR-122-5pNon-templated addition 3’ A LysCTT HisGTG ArgCCG SNORA9 5’ 5’ 5’ 5’ 5’3’ 3’ 3’ 3’ 3’ •Tissue-of-origin of plasma DNA from a healthy individual deduced from analysis of nucleosome spacing signals downstream of transcription start sites for ssDNA-seq1 and TGIRT-seq • Plasma RNA fragments from diverse protein-coding genes • Full-length sncRNAs • Differentially represented transcripts identified from plasma vesicle RNA- seq correlated with survival based on microarray data • mRNAs may be cell- type-specific exosome cargos • Comparisons to TruSeq v2 and v3 for ribo-depleted, fragmented human reference RNA samples plus ERCC spike-ins1 • More 5’ proximal splice junctions and embedded snoRNAs 1) RNA or DNA fragments 3’5’ 3’5’ 3) Total RNA/DNA method 3) Small RNA method 3’N 3’ blocker RNA-seq adapter TGIRT 5’ 2) Template-switching DNA primer 5’ 5’ 5’ App 3’ 3’ 3’ blocker Target RNA cDNA Target RNA cDNA 4) PCR amplification Target RNA cDNA Target RNA P7P5 5’ 3’ 3’ 5’ ~1 ng 20 min 60 min 1. Snyder, et al. Cell 2016 Wu and Lambowitz. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Scientific Reports 2017