Defining the relevant genome in solid tumors
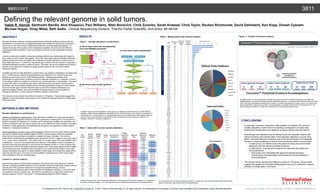
- 1. Habib R. Hamidi, Santhoshi Bandla, Nick Khazanov, Paul Williams, Nikki Bonevich, Chris Zurenko, Sarah Anstead, Chris Taylor, Reuban Richmonds, David Galimberti, Ken Kopp, Dinesh Cyanam, Michael Hogan, Vinay Mittal, Seth Sadis. Clinical Sequencing Division, Thermo Fisher Scientific, Ann Arbor, MI 48104 RESULTSABSTRACT Decades of cancer research including comprehensive molecular profiling combined with the development of a broad array of targeted therapies have created the opportunity to transform cancer care in the near future by implementing precision oncology based approaches. An important element of this system is the widespread availability of robust and cost-effective multivariate profiling methods in order to characterize relevant cancer associated molecular alterations. Current commercially available multivariate profiling methods vary dramatically with regard to the number of cancer genes interrogated. Given that many large scale and detailed molecular profiling studies have been completed, the landscape of somatic alterations in solid tumors is reasonably well-known. Furthermore, the specific gene variants that are relevant to application of targeted therapies are also a matter of record. Therefore, we set out to define the number of relevant cancer genes for precision oncology research based on the currently available empirical evidence. To define recurrent somatic alterations in solid tumors, we created a compendium of variant calls from >10,000 exomes, defined focal amplifications and deletions from >30,000 arrays, and defined recurrent fusions from >8,000 RNAseq profiles. Statistical approaches were implemented to define genes containing recurrent missense mutations (i.e., hotspots), enriched in truncating mutations or subject to recurrent copy number gain/loss or translocation. This gene set was then used to comprehensively search approved cancer drug labels, clinical practice guidelines, and clinical trials to identify records containing published evidence in which specific recurrent somatic gene variants that were used as part of the indication statement of an approved targeted therapy, were recommended for testing as part of clinical practice for therapeutic decisions, or were used as enrollment criteria in clinical trials. The relevant cancer genome thus defined consists of <100 genes. These results suggest that targeted multivariate profiling approaches may be sufficient to support precision oncology goals in the near future. MATERIALS AND METHODS Somatic alterations in solid tumors. Identifying significantly mutated genes. Data obtained from different sources were annotated using an annotation pipeline optimized to identify significantly mutated genes. The Oncomine™ method compared the frequency of mutations with a background mutation rate estimation. The Fisher’s combination test was used to assess the overall significance of the mutation in the gene of interest. It tests against the null hypothesis that the gene is not significant in either hotspot mutations or deleterious mutations. Genes subjected to recurrent copy number gains/loss. Minimal common region (MCR) analysis was used to identify focally amplified regions. Briefly, common regions (CR) were identified which are chromosomal regions amplified or deleted in at least 2 samples. The minimum thresholds for amplifications and deletions were set at log2 ≥ 0.9 (3.7 copies or more) and log2 ≤ -0.9 (1 copy or less) respectively. Then within each CR, a peak region (PR) was identified which is defined as— (i) one or more genes that were aberrant in the highest number of samples (n) and also those that were aberrant in one less than the highest number (n-1) and (ii) genes that were aberrant in 90% of the highest aberrant sample count. These peak regions across multiple cancer types were then clustered using Cytoscape 2.8.3 to build network clusters. The cluster analysis compared every gene in a peak to genes in other peaks and clusters the peaks with at least one common gene. The most recurrent amplified or deleted gene(s) within each cluster was considered a recurrent candidate gene Curation of relevant evidence. Approved drug labels, clinical practice guidelines, and clinical trials were searched to identify records containing published evidence in which specific recurrent somatic gene variants were (i) used as part of the indication statement of an approved targeted therapy, (ii) were recommended for testing as part of clinical practice for therapeutic decisions, or (iii) were used as enrollment criteria in clinical trials. All content is compiled and curated each quarter and reported using Ion Torrent™ Oncomine™ Knowledgebase Reporter (OKR) software. CONCLUSIONS • A systematic evaluation of genomics data enabled us to identify 476 recurrent somatic alterations in solid tumors by creating a compendium of variant calls, defined focal amplifications and deletions as well as defined recurrent fusions. • Relevant genomic alterations can be defined as the set of genetic variants with relevant evidence documented either in the indication statement in cancer drug labels, recommendations for testing as part of the clinical practice for therapeutic decision in clinical guidelines or as part of the enrollment criteria of clinical trials. • In solid tumors, we defined a set of 82 genes as having recurrent somatic alterations that had relevant published evidence. • In solid tumors, 12 genes were supported by approved drug labels and clinical guidelines. • Nine genes were associated with approved targeted therapies. • Twelve genes were associated with therapeutic recommendations in clinical guidelines. • The relevant cancer genome thus defined consists of <100 genes. These results suggest that targeted multivariate profiling approaches may be sufficient to support precision oncology goals in the future. Defining the relevant genome in solid tumors. For Research Use Only. Not for use in diagnostic procedures. © 2017 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Thermo Fisher Scientific • 5781 Van Allen Way • Carlsbad, CA 92008 • thermofisher.com Gene Symbol Mutated Sample Frequency Mutated Sample Count Sample Count Oncomine Gene Classification Hotspot Frequency in Mutated Samples Hotspot Q Value Deleterious Frequency in Mutated Samples Deleterious Q Value TP53 29.90% 3052 10194 Loss of Function 64.30% 1.20E-14 26.70% 0.00E+00 PIK3CA 10.30% 1049 10194 Gain of Function 90.00% 1.20E-14 1.00% 1.00E+00 PCLO 8.00% 815 10194 Gain of Function 12.00% 4.80E-11 13.60% 7.60E-01 KRAS 6.50% 659 10194 Gain of Function 96.20% 1.20E-14 0.00% 1.00E+00 KMT2C 6.20% 627 10194 Loss of Function 5.10% 1.10E-01 33.70% 0.00E+00 BRAF 6.10% 618 10194 Gain of Function 84.00% 7.40E-12 1.00% 1.00E+00 MUC4 5.70% 577 10194 Gain of Function 29.50% 1.20E-14 4.20% 1.00E+00 KMT2D 5.40% 553 10194 Loss of Function 2.00% 1.40E-03 39.10% 0.00E+00 PTEN 5.40% 552 10194 Loss of Function 27.50% 1.20E-14 49.80% 0.00E+00 ARID1A 5.30% 542 10194 Loss of Function 4.40% 9.10E-11 65.50% 0.00E+00 APC 5.20% 535 10194 Loss of Function 1.10% 7.10E-01 65.00% 0.00E+00 FAT1 4.40% 453 10194 Loss of Function 1.30% 8.70E-01 35.10% 0.00E+00 CTNNB1 4.20% 432 10194 Gain of Function 79.40% 1.20E-14 3.70% 1.00E+00 IDH1 4.20% 425 10194 Gain of Function 90.10% 1.20E-14 0.90% 1.00E+00 NF1 3.80% 391 10194 Loss of Function 1.80% 3.80E-02 42.70% 0.00E+00 ATRX 3.80% 384 10194 Loss of Function 5.50% 1.30E-11 37.20% 0.00E+00 SACS 3.70% 375 10194 Loss of Function 6.10% 2.50E-03 17.90% 1.30E-05 ATM 3.60% 372 10194 Loss of Function 13.70% 1.60E-11 22.30% 2.40E-12 ZFHX3 3.60% 369 10194 Loss of Function 10.30% 1.20E-14 23.30% 2.70E-14 FMN2 3.60% 365 10194 Gain of Function 21.10% 1.20E-14 6.80% 1.00E+00 A total of 476 genes had recurrent somatic alterations including recurrent missense mutations (i.e. hotspots) or enriched in truncating mutation. The top 20 genes based on mutated sample frequency are displayed above. Table 1. Genes with recurrent somatic alterations. Table 2. Mutated genes with relevant evidence. As of December 2016, 82 genes contained evidence in at least one of the three clinically relevant data sources (global clinical trials, approved label and clinical guidelines). Figure 2. Curation of relevant evidence. Identified records in which specific gene variants were used as part of the indication statement of an approved targeted therapy, were part of clinical practice for therapeutic decisions, or were used as enrollment criteria in clinical trials. Data was systematically assessed and curated for evidence associated with a gene, variant, and cancer type that was specified in approved drug labels, clinical guidelines, and global clinical trials. As of December 2016, 82 genes contained evidence in at least one of these three clinically relevant data sources. Gene Number of Trials Number of Approved Labels Number of Clinical Guidelines Total Evidence Count ERBB2 253 8 6 267 EGFR 250 9 3 262 BRAF 103 8 5 116 KRAS 80 4 5 89 ALK 78 5 3 86 NRAS 40 4 3 47 MET 39 0 1 40 PIK3CA 35 0 0 35 KIT 26 0 4 30 FGFR1 24 0 0 24 HRAS 24 0 0 24 PTEN 24 0 0 24 FGFR2 22 0 0 22 FGFR3 22 0 0 22 PDGFRA 20 0 2 22 ROS1 17 2 1 20 TP53 18 0 0 18 AKT1 17 0 0 17 MTOR 16 0 0 16 RET 15 0 1 16 TSC1 15 0 0 15 TSC2 15 0 0 15 FGFR4 12 0 0 12 AKT3 11 0 0 11 NTRK1 11 0 0 11 PIK3R1 11 0 0 11 CD274 10 0 0 10 NTRK3 10 0 0 10 RHEB 10 0 0 10 STK11 10 0 0 10 NTRK2 10 0 0 10 BRCA1 8 1 0 9 BRCA2 8 1 0 9 CDKN2A 8 0 0 8 IDH1 7 0 0 7 KDR 6 0 0 6 NF1 5 0 1 6 RAF1 6 0 0 6 CDK4 5 0 0 5 ERBB3 5 0 0 5 GNA11 5 0 0 5 GNAQ 5 0 0 5 MAP2K1 5 0 0 5 MAP2K2 5 0 0 5 NF2 5 0 0 5 ARAF 4 0 0 4 CCND1 4 0 0 4 MAPK1 4 0 0 4 PTCH1 4 0 0 4 ABL1 3 0 0 3 AR 3 0 0 3 CDK6 3 0 0 3 DDR2 3 0 0 3 FLT3 3 0 0 3 GNAS 3 0 0 3 IDH2 3 0 0 3 PTPN11 3 0 0 3 SMO 3 0 0 3 ATM 2 0 0 2 AXL 2 0 0 2 BAP1 2 0 0 2 CSF1R 2 0 0 2 ESR1 2 0 0 2 JAK2 2 0 0 2 MYC 2 0 0 2 RB1 2 0 0 2 SMARCB1 2 0 0 2 CBL 1 0 0 1 CCNE1 1 0 0 1 CHEK2 1 0 0 1 ERBB4 1 0 0 1 ERG 1 0 0 1 FBXW7 1 0 0 1 FOXL2 1 0 0 1 JAK1 1 0 0 1 JAK3 1 0 0 1 MDM2 1 0 0 1 MYCN 1 0 0 1 NOTCH1 1 0 0 1 PPARG 1 0 0 1 SRC 1 0 0 1 STAT3 1 0 0 1 Figure 1. Somatic alterations in solid tumors. To define recurrent somatic alterations in solid tumors, we defined recurrent fusions from >8,000 RNAseq profiles(a), defined focal amplifications and deletions from >30,000 arrays(b), and created a compendium of variant calls from >15,000 exomes (c). Statistical approaches were implemented to define genes subject to recurrent copy number gain/loss or translocations (b); or containing recurrent missense mutations (i.e., hotspots), enriched in truncating mutations (c). (a) Novel fusion calls with corresponding exon-level RNASeq expression. (b) Recurrent copy number gain/loss. (c) Flow chart of gene classification. 3811 Genes(N=31)withmorethan10totalevidencesources. Total Evidence Count
