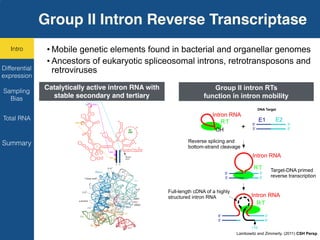
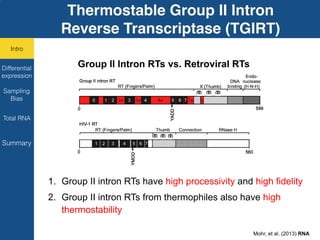
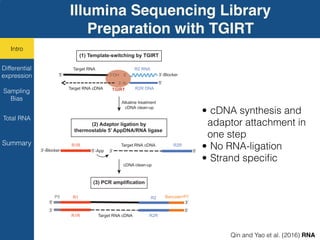
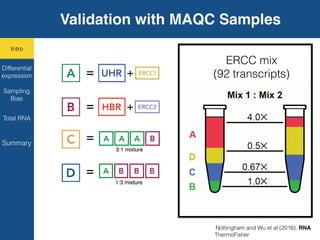
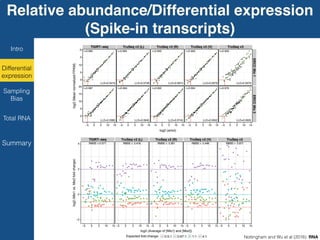
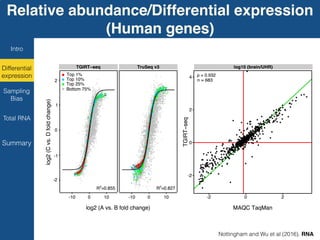
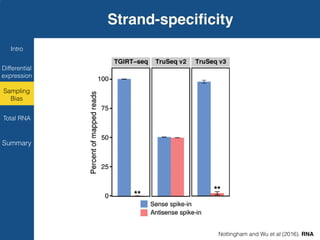
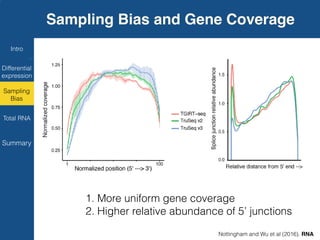
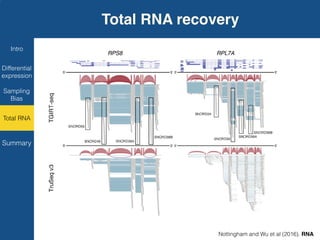
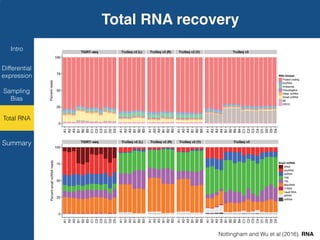
TGIRT is a thermostable group II intron reverse transcriptase that enables high-throughput RNA sequencing with several advantages over other reverse transcriptases. It recapitulates the relative abundance of transcripts, has higher strand-specificity, and provides more uniform gene coverage from 5' to 3' compared to TruSeq v3. TGIRT also enables simultaneous profiling of mRNA, lncRNA, and other structured small RNAs. Its applications include structured RNA-seq, plasma and exosomal RNA-seq for cancer diagnostics, and generation of long cDNAs.