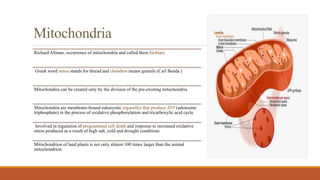
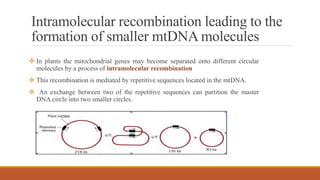
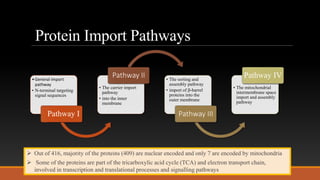
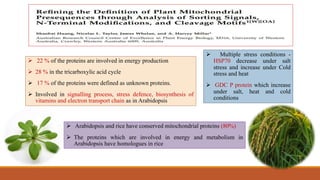
Mitochondria are membrane-bound organelles found in eukaryotic cells that produce ATP through oxidative phosphorylation. Plant mitochondria contain their own circular DNA and can divide through binary fission. Mitochondrial genes are regulated through transcription, splicing, editing, and post-transcriptional mechanisms. The mitochondrial proteome is comprised mostly of nuclear-encoded proteins that are imported through protein transport pathways. Mitochondria play key roles in energy production, reactive oxygen species regulation, and programmed cell death signaling in plants. Mitochondrial omics research can provide insights into stress responses and enhance stress tolerance in plants.