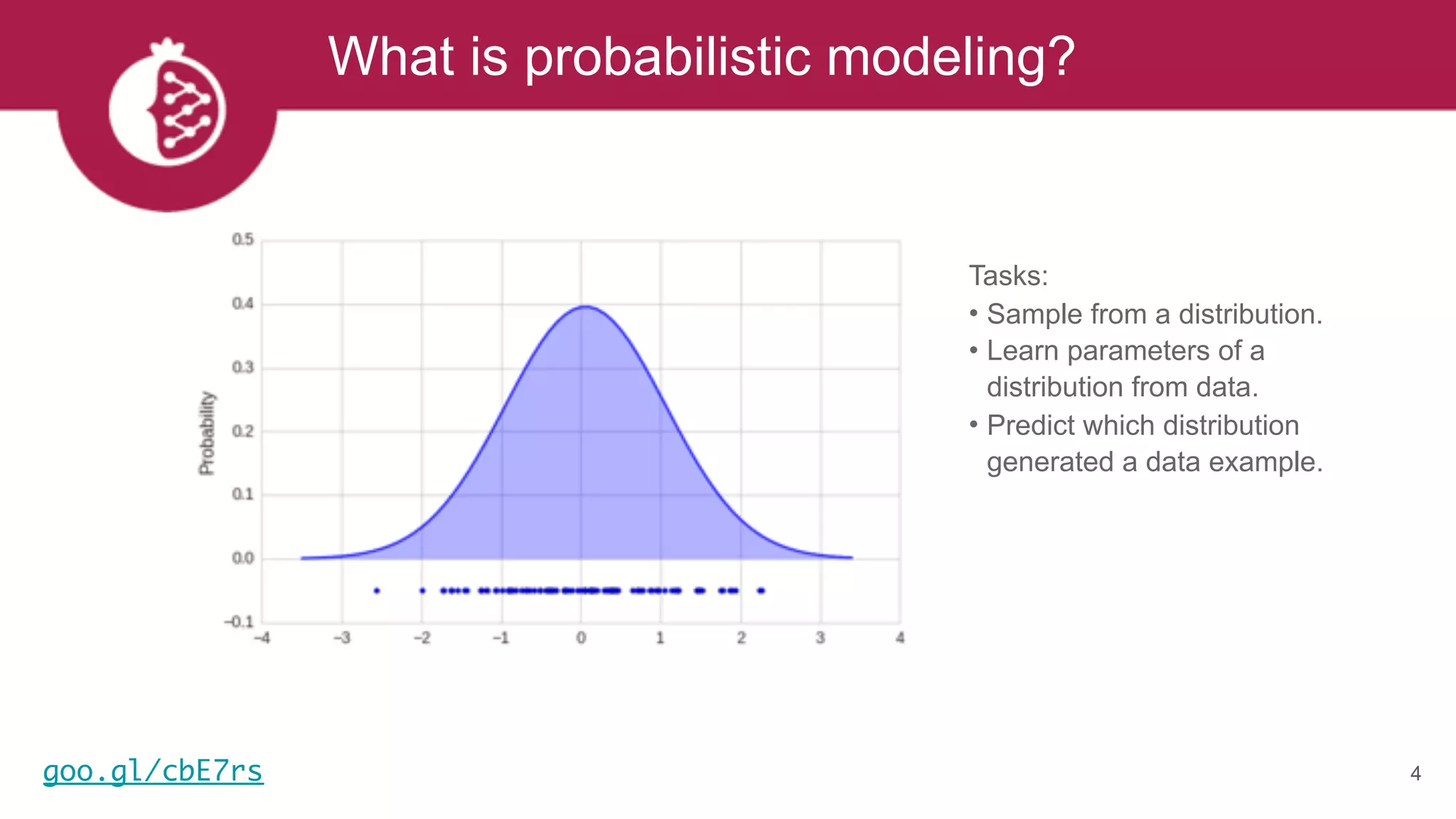
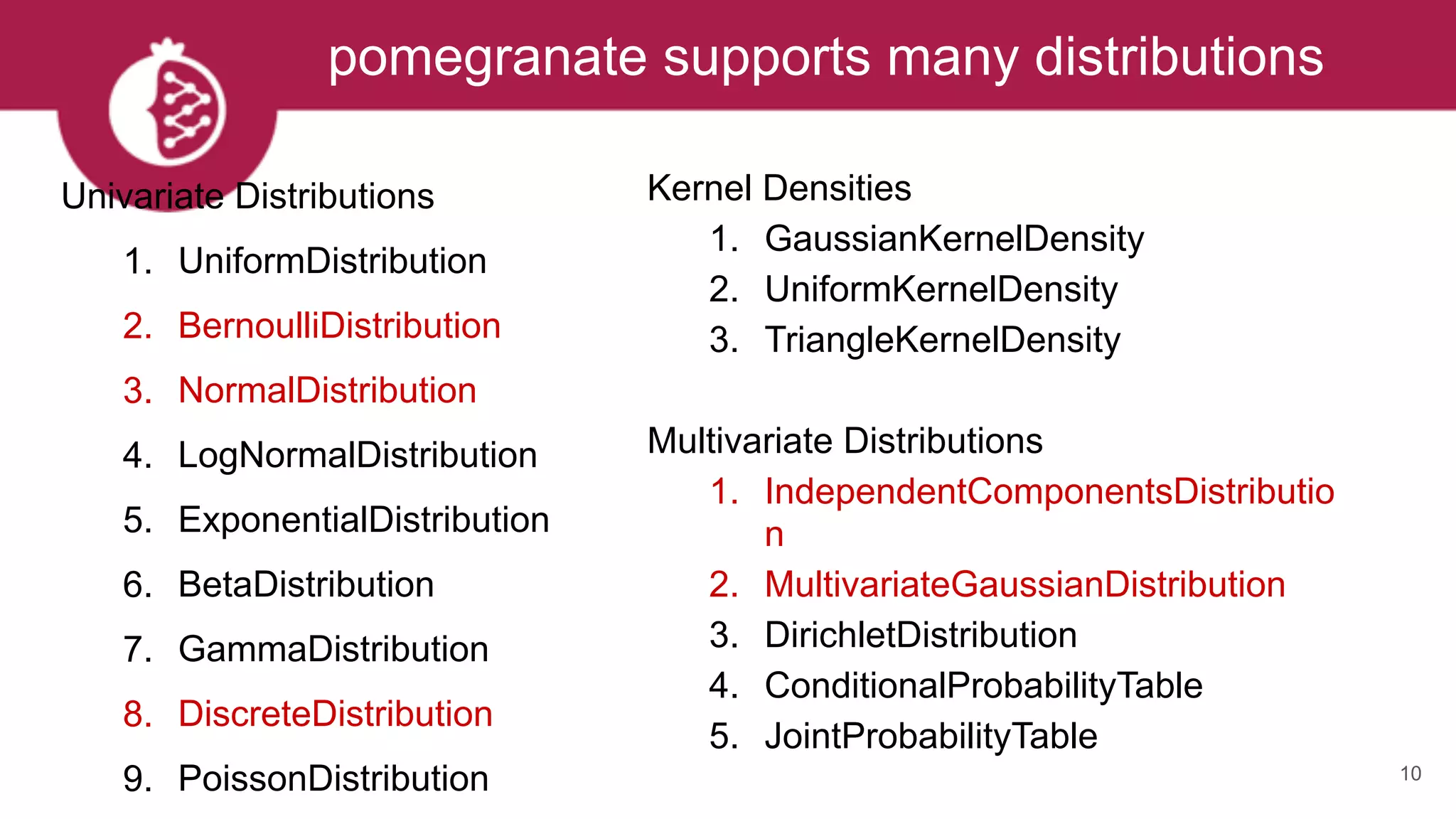
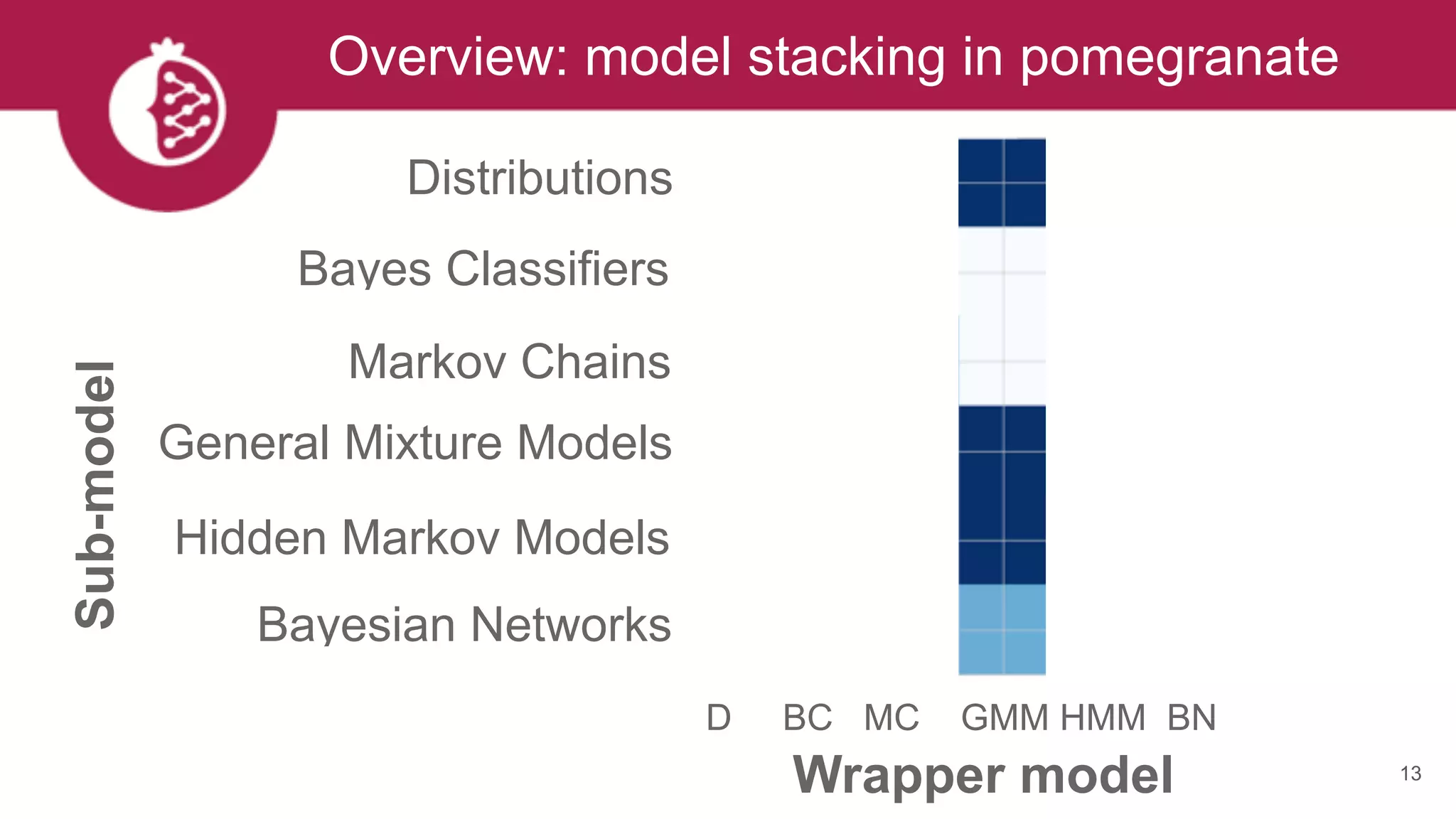
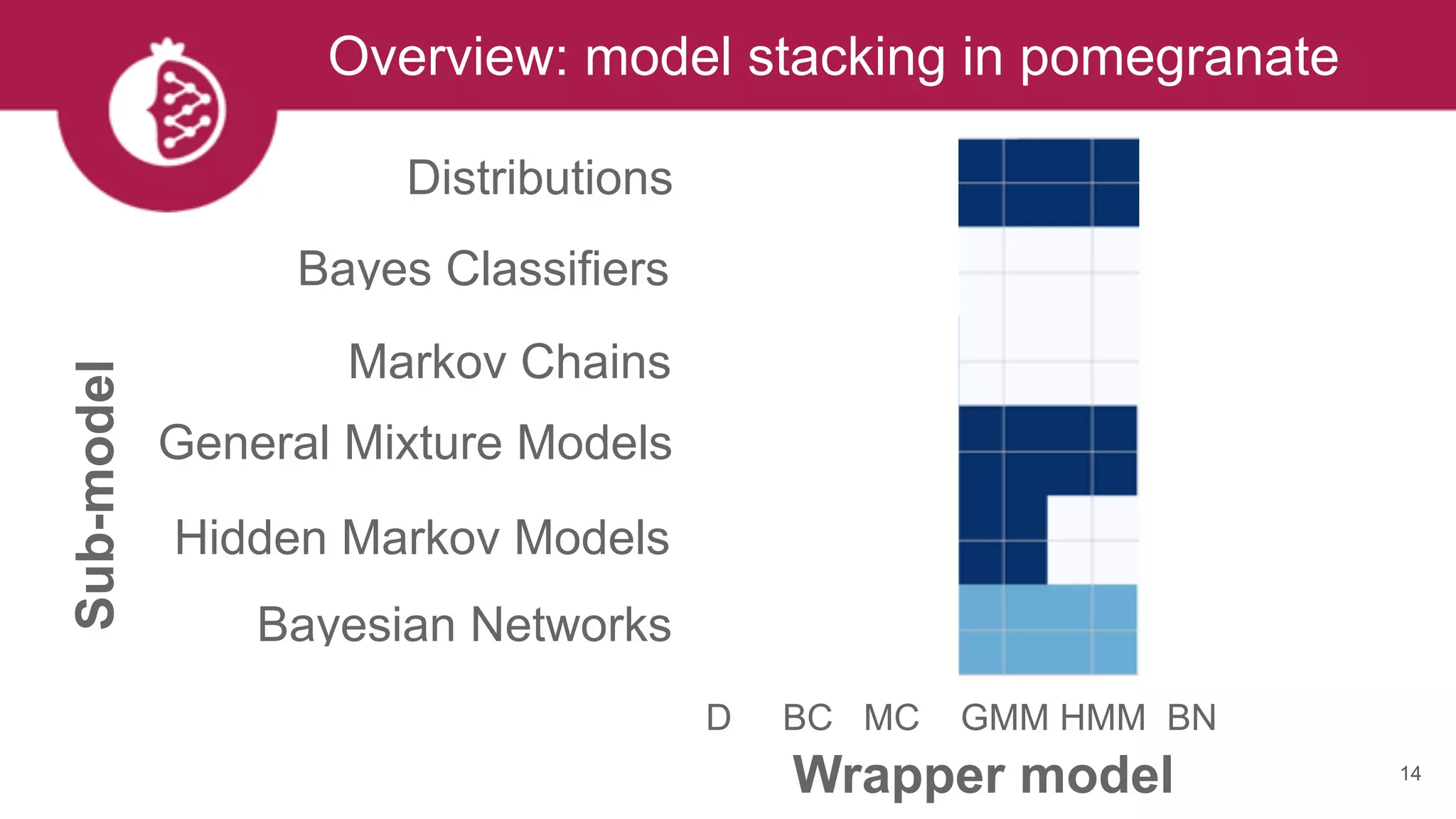
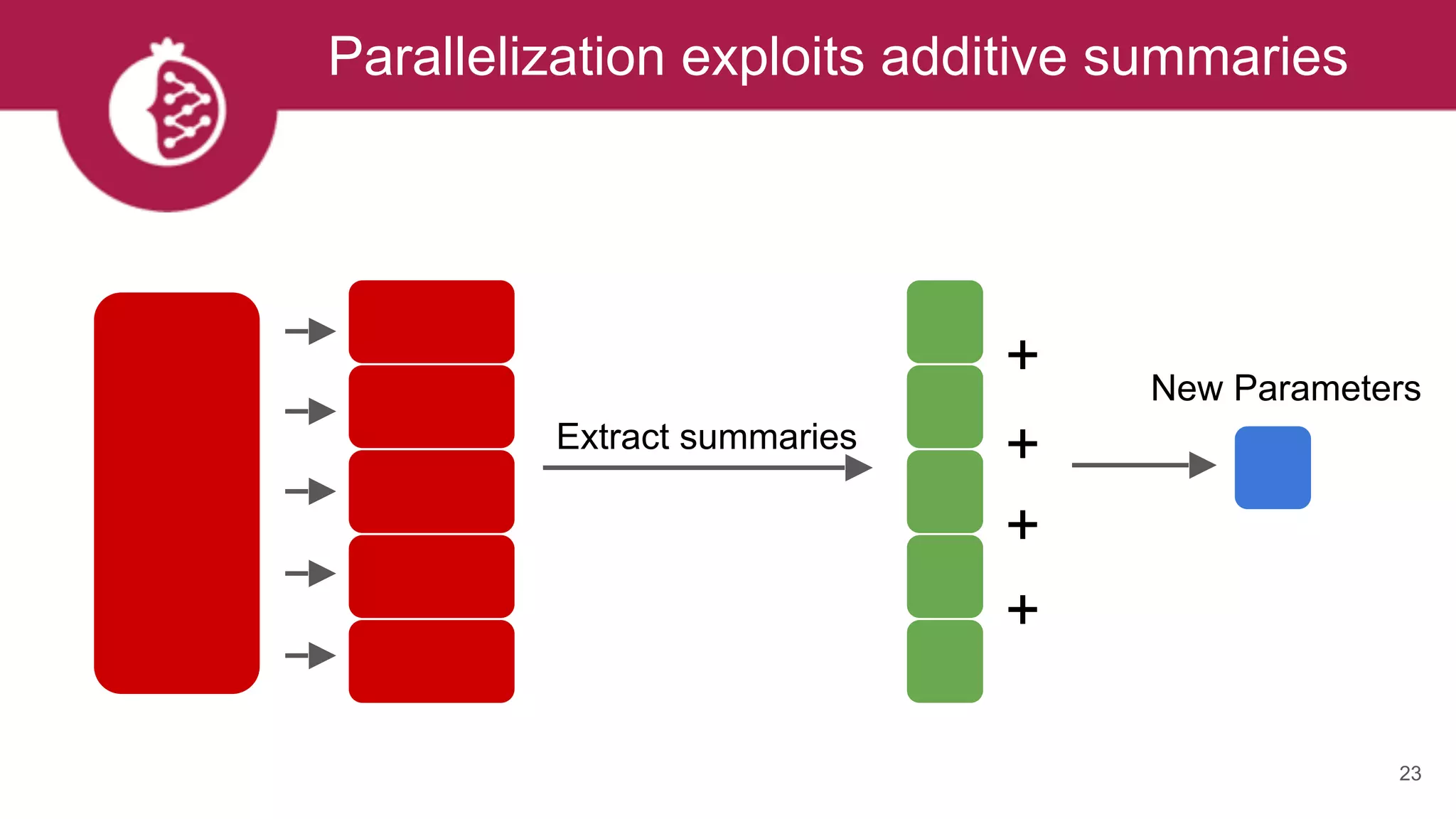
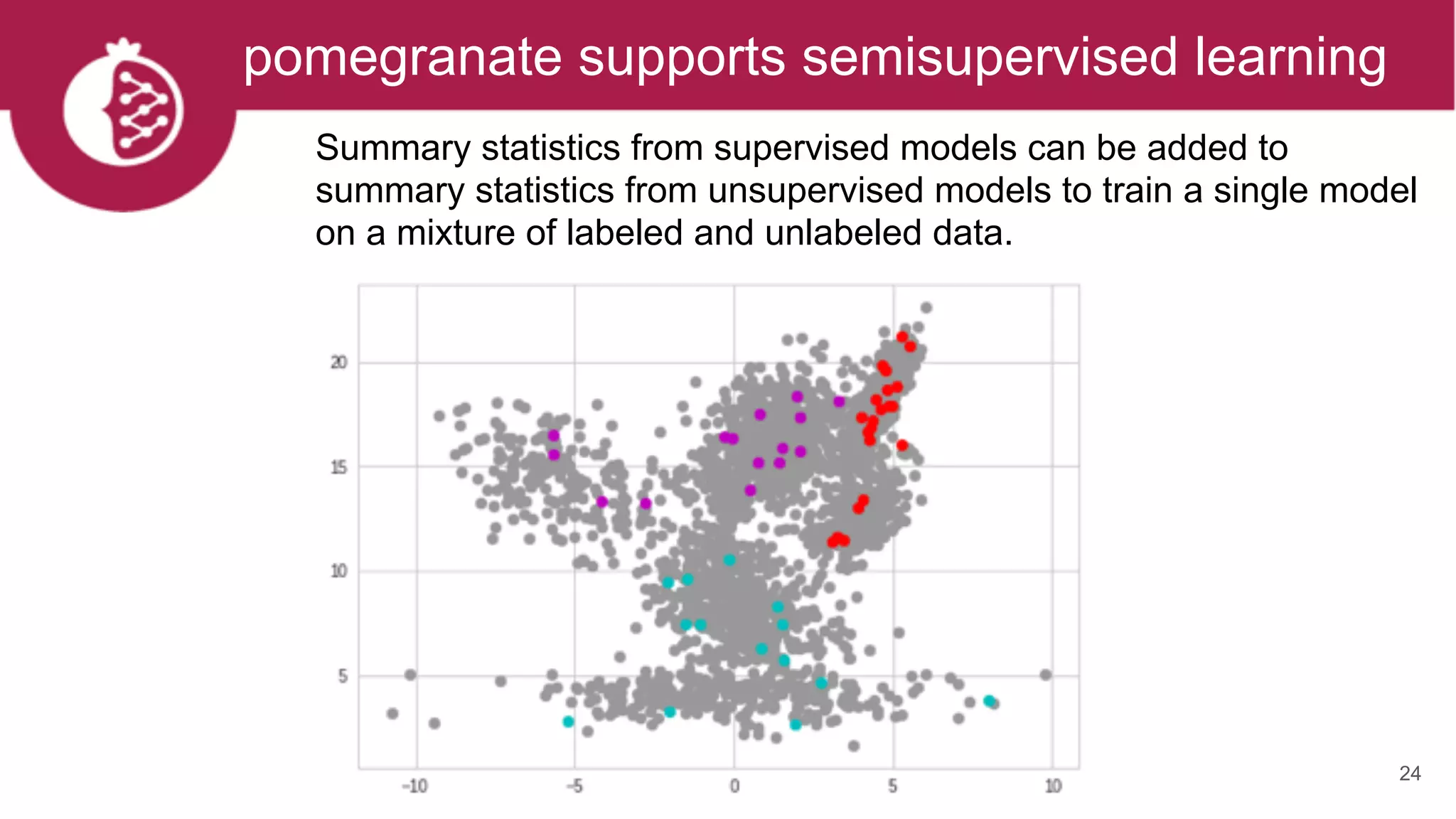
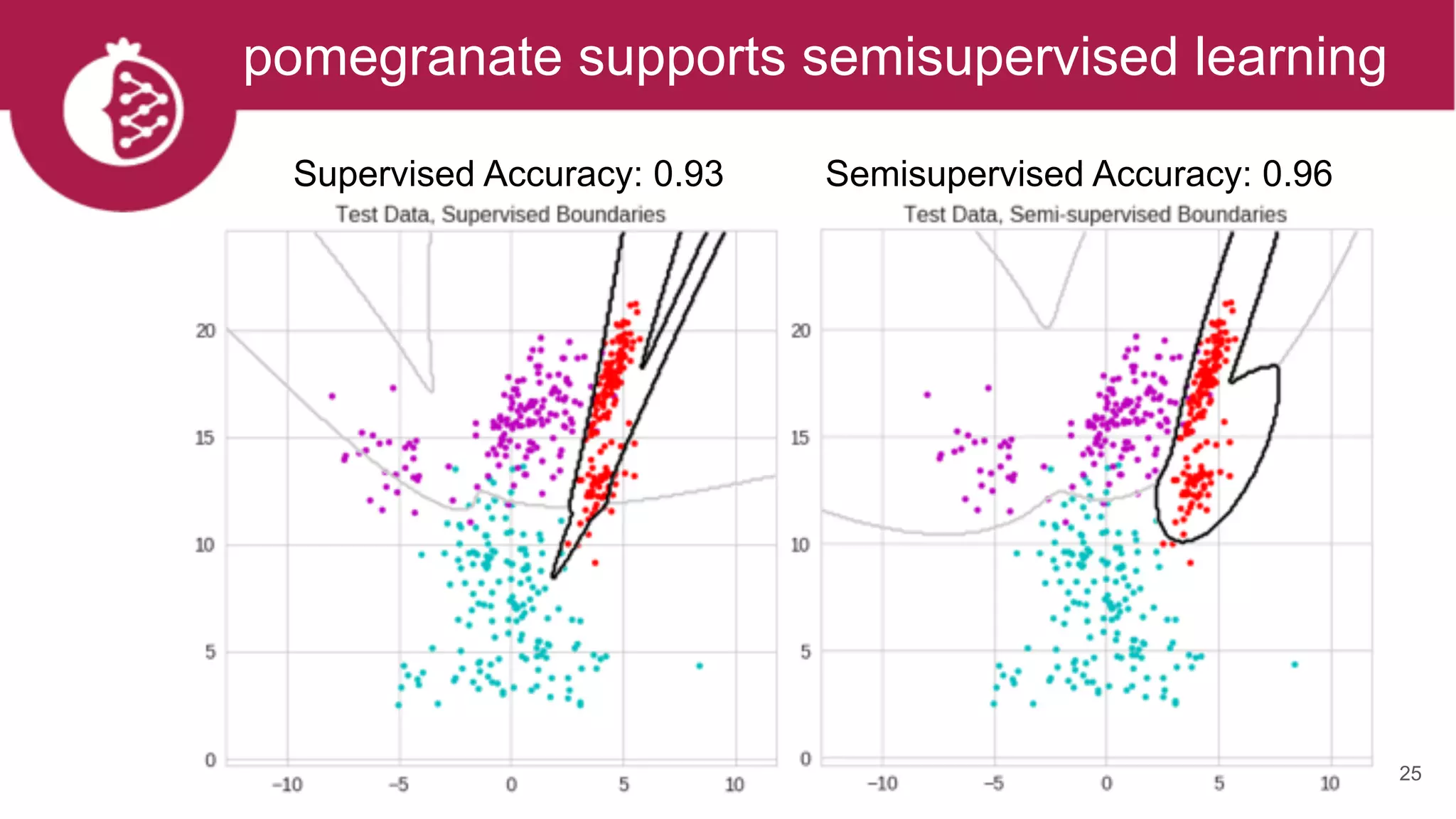
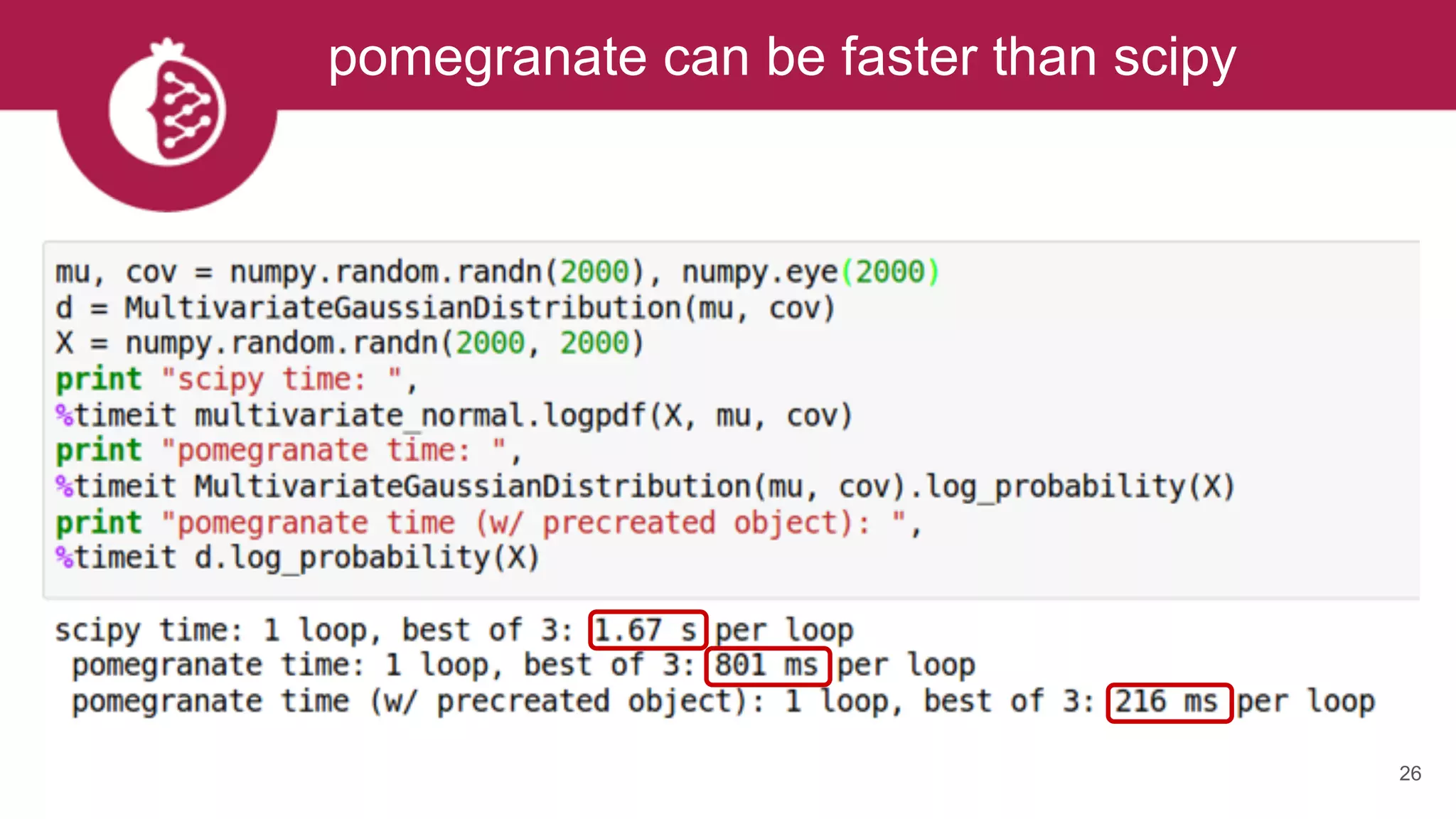
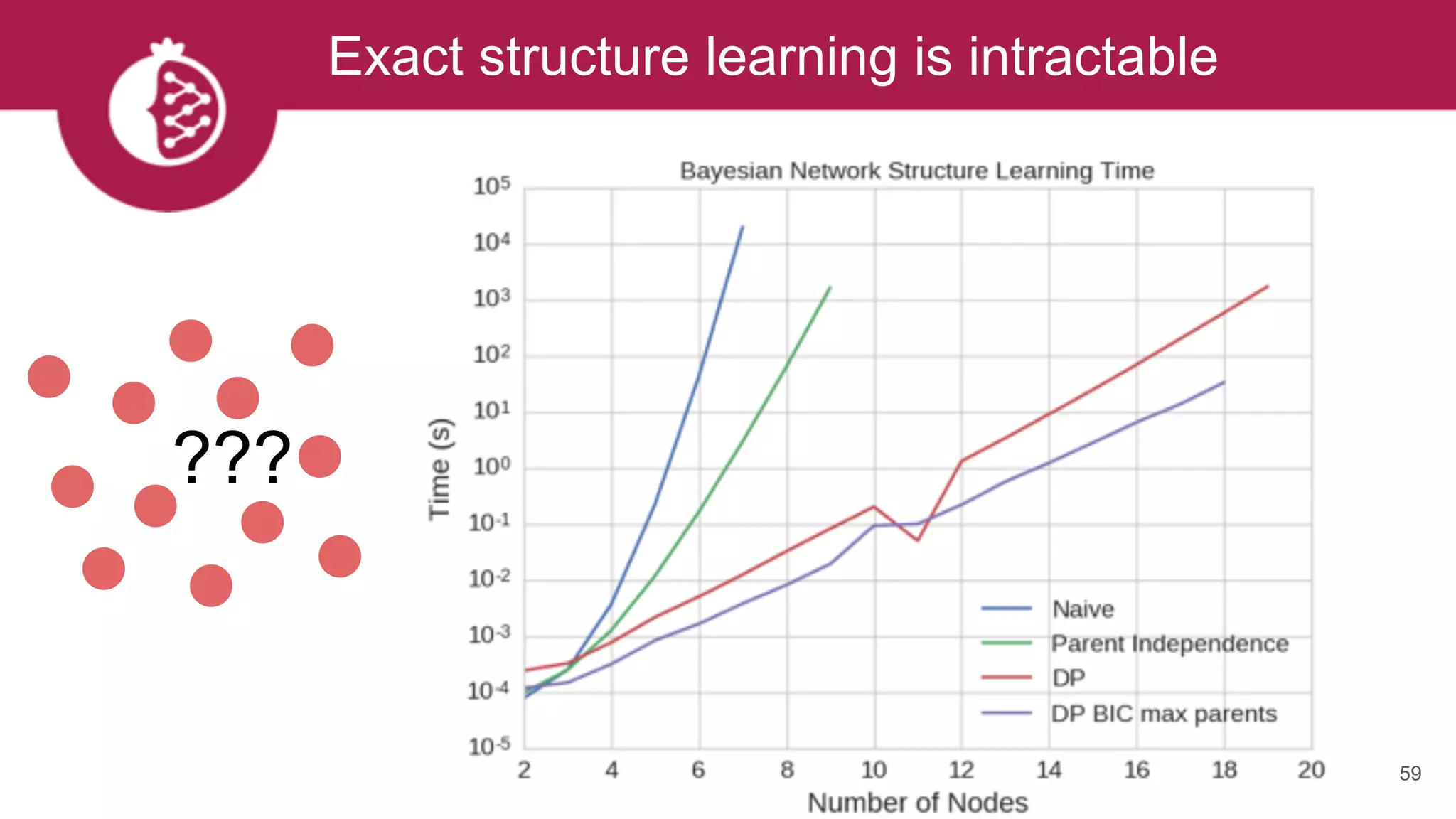
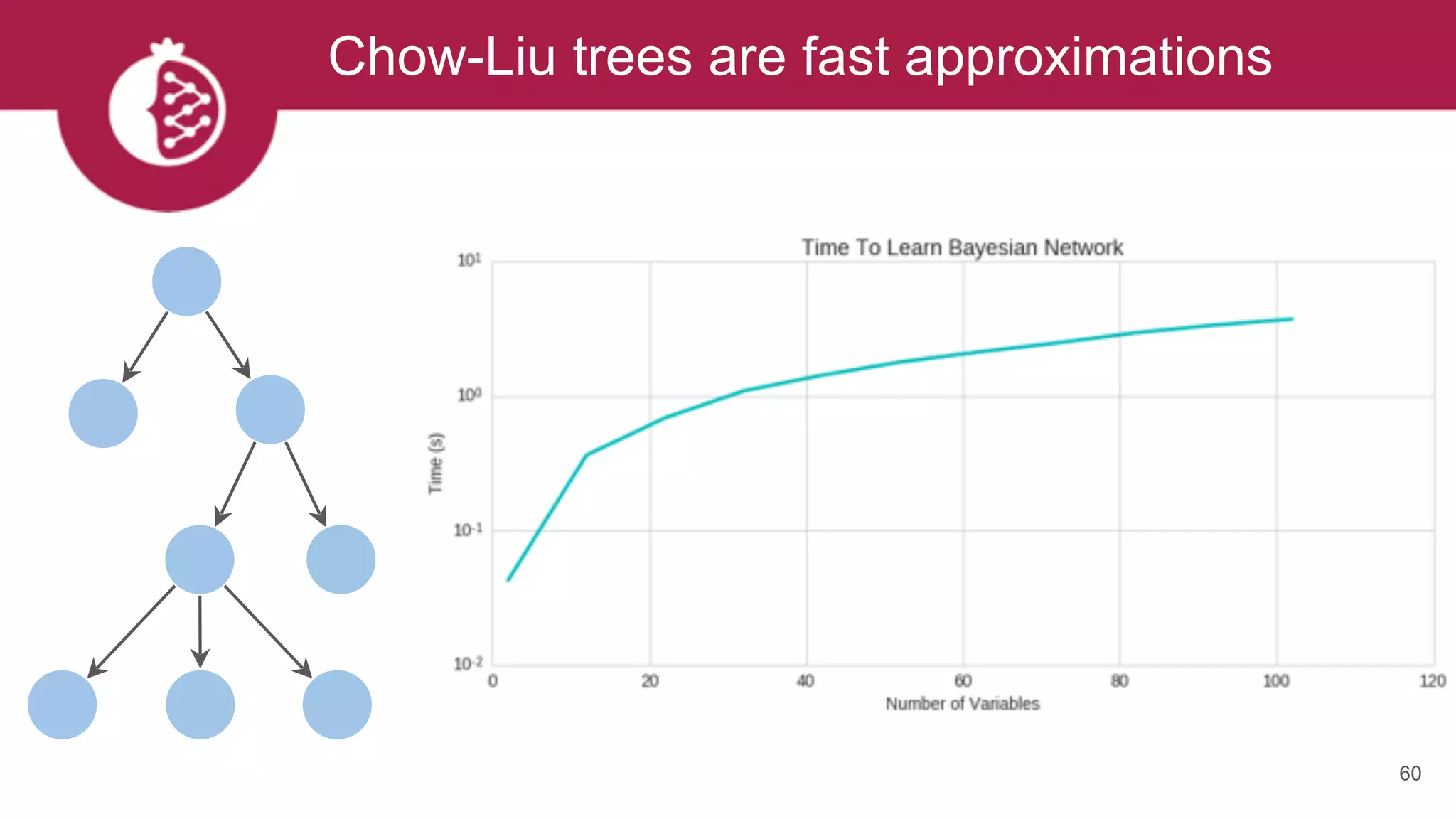
This document discusses probabilistic modeling in Python using the pomegranate library. It begins with an overview of probabilistic modeling tasks and when to use pomegranate. It then discusses the major models supported by pomegranate including general mixture models, hidden Markov models, Bayesian networks, and Bayes classifiers. It provides examples of using these models and shows how pomegranate allows for complex probabilistic models to be built from simpler component distributions and submodels. The document concludes by demonstrating how mixtures of hidden Markov models can be trained in parallel using pomegranate.

![11
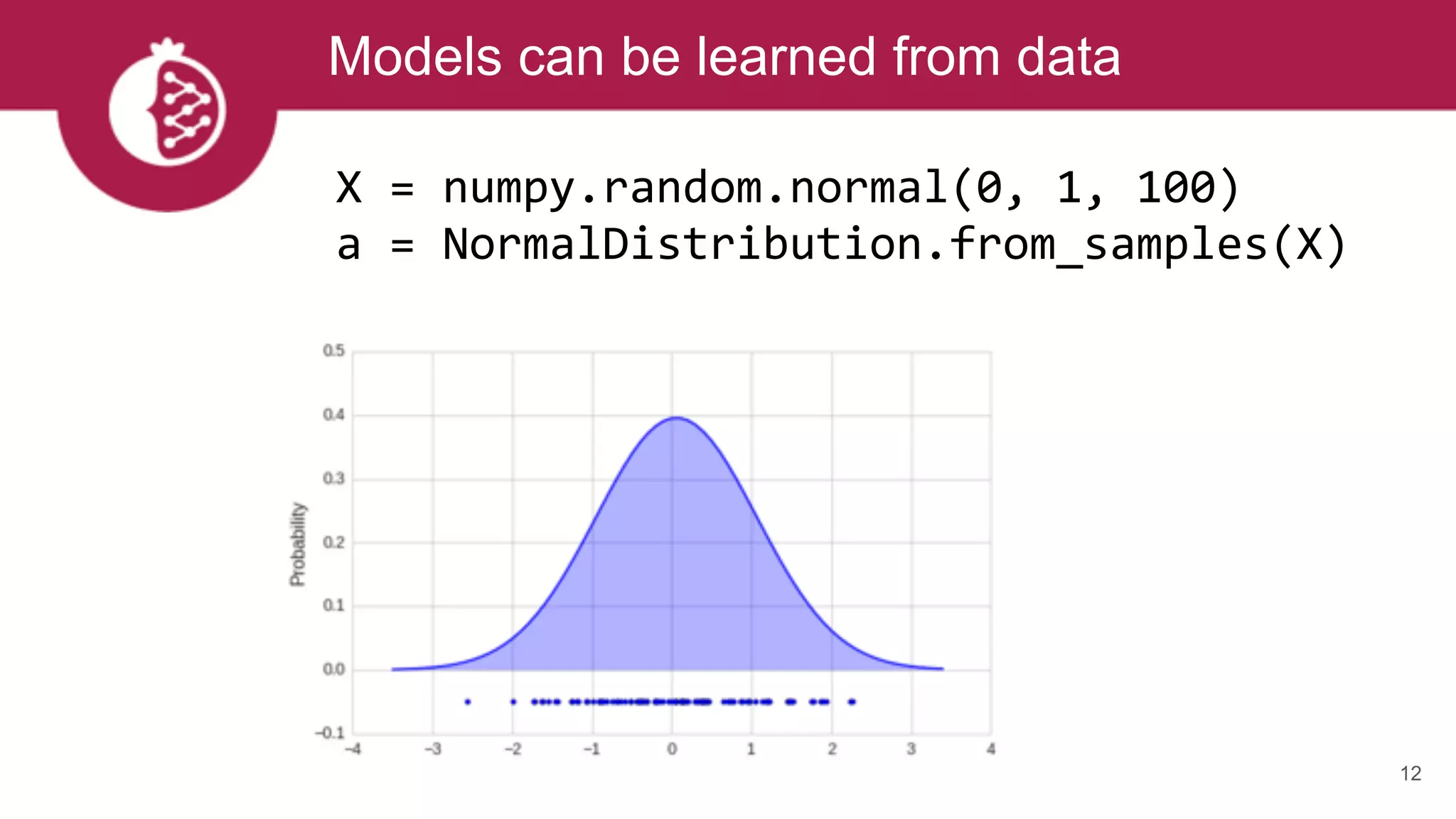
mu, sig = 0, 2
a = NormalDistribution(mu, sig)
X = [0, 1, 1, 2, 1.5, 6, 7, 8, 7]
a = GaussianKernelDensity(X)
Models can be created from known values](https://image.slidesharecdn.com/tzxq8ddyrpomx9xvfny5-signature-a56a2239ee4119c26e3e070c0ece32f2898146a45b6997205cf22bd9d4505024-poli-170721202738/75/Maxwell-W-Libbrecht-pomegranate-fast-and-flexible-probabilistic-modeling-in-python-11-2048.jpg)
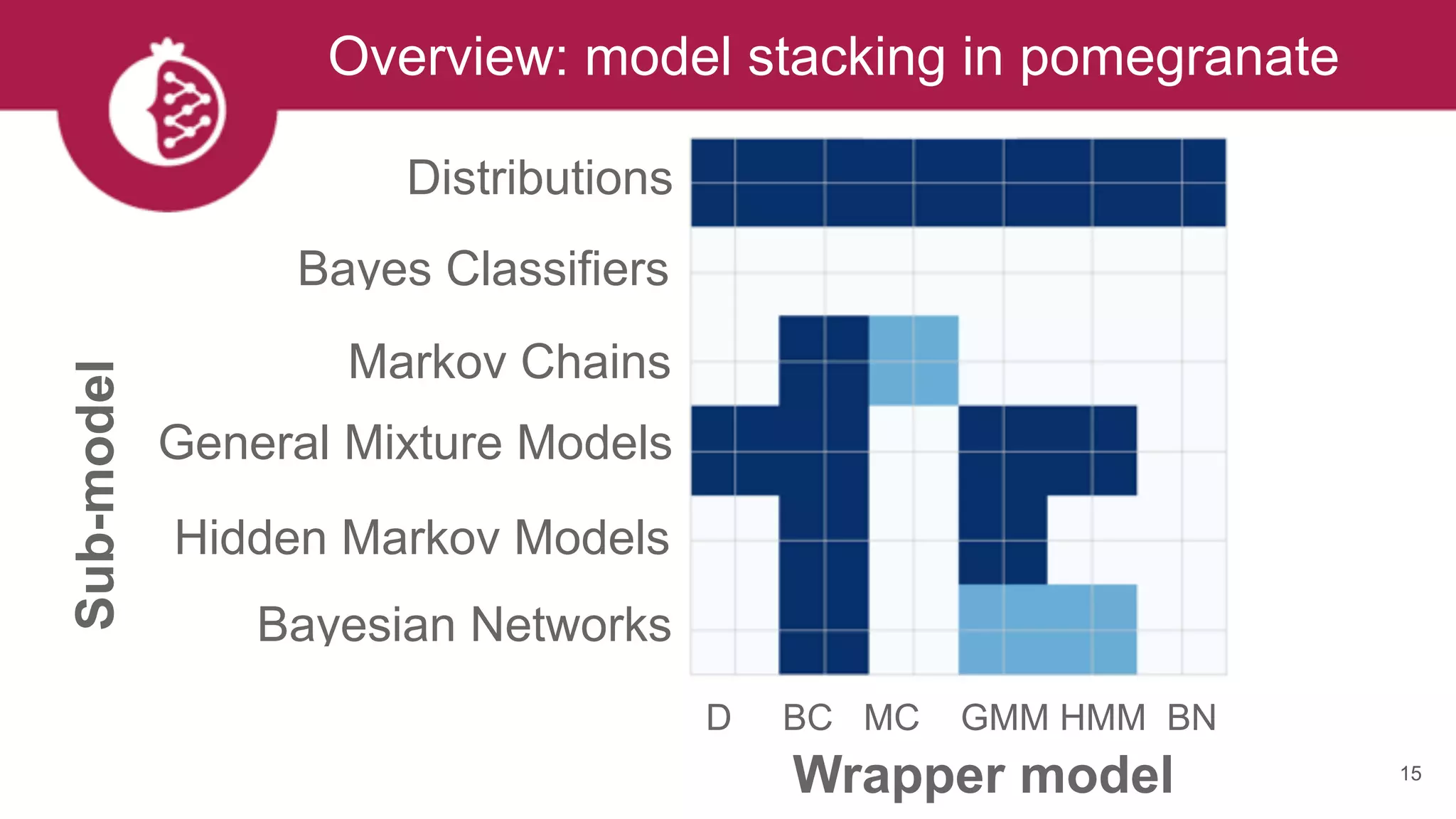
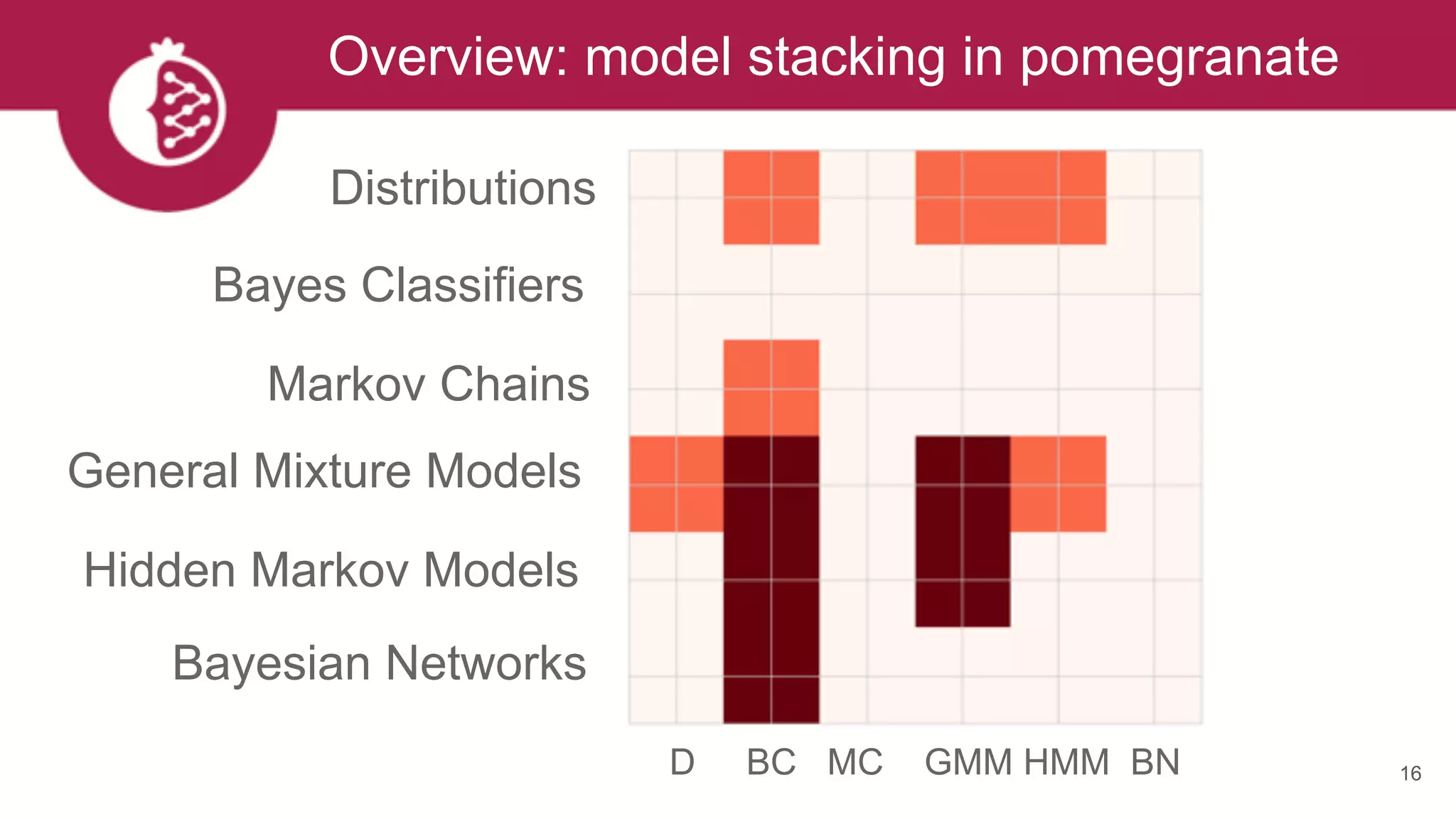
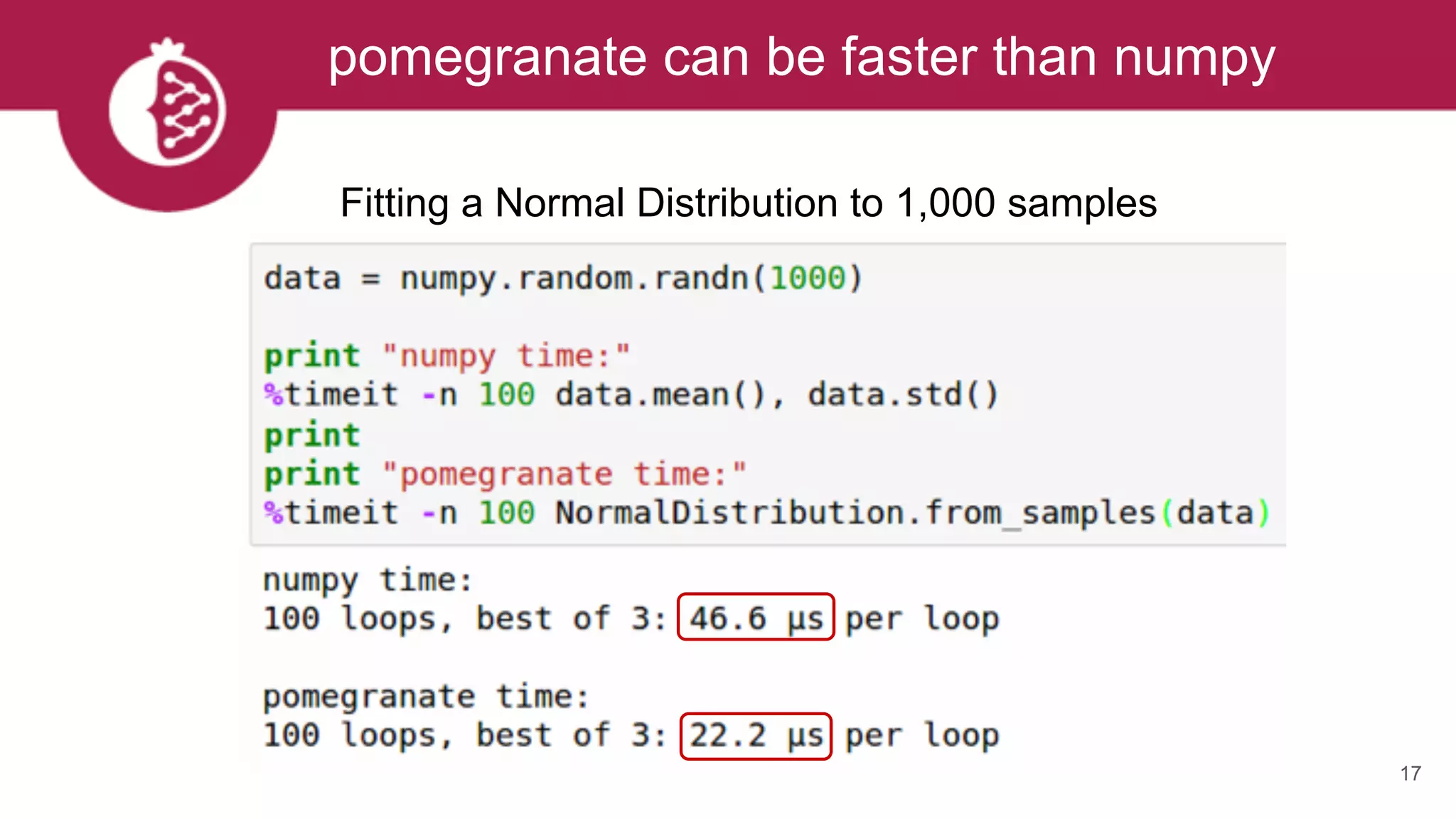
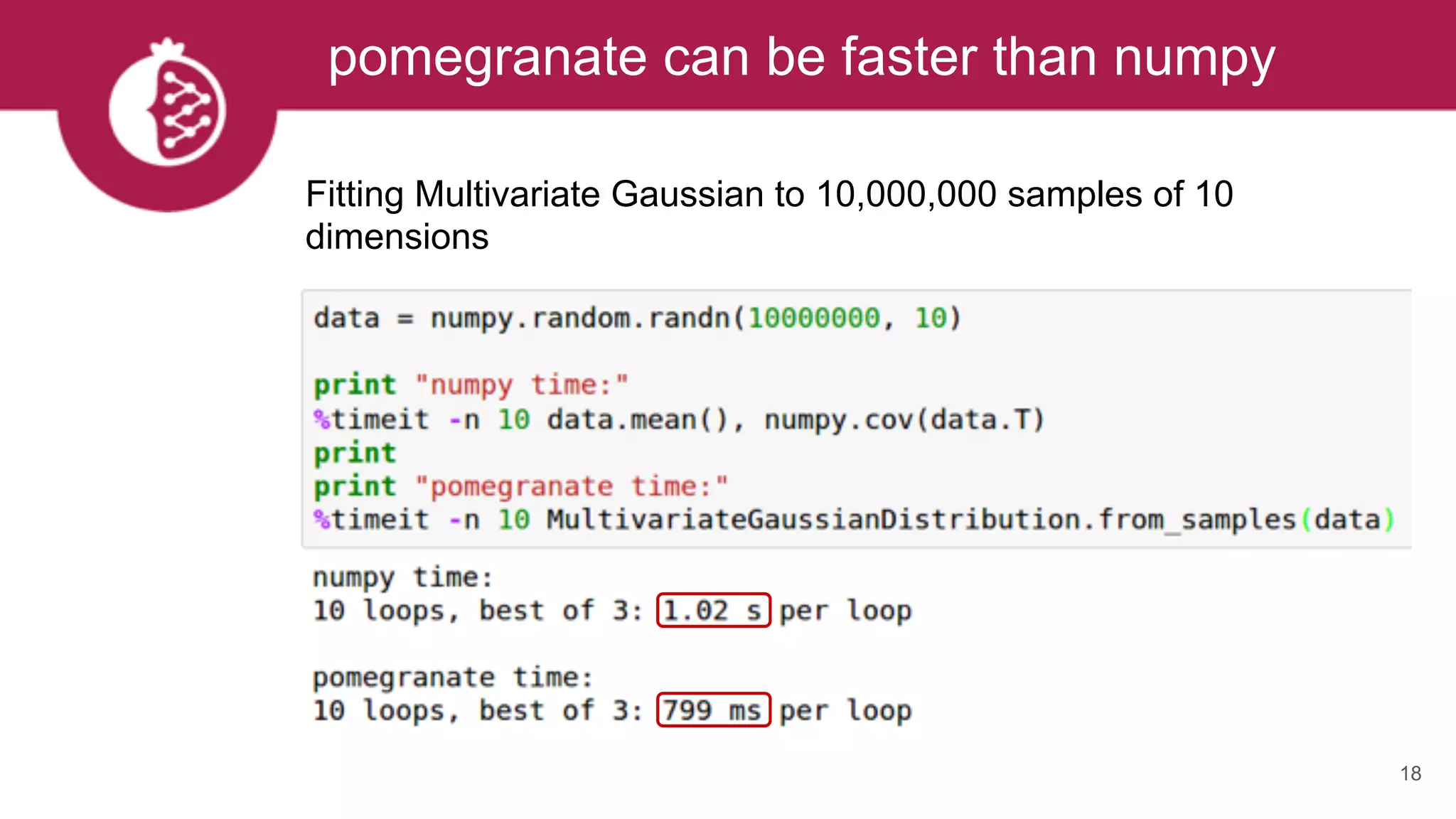





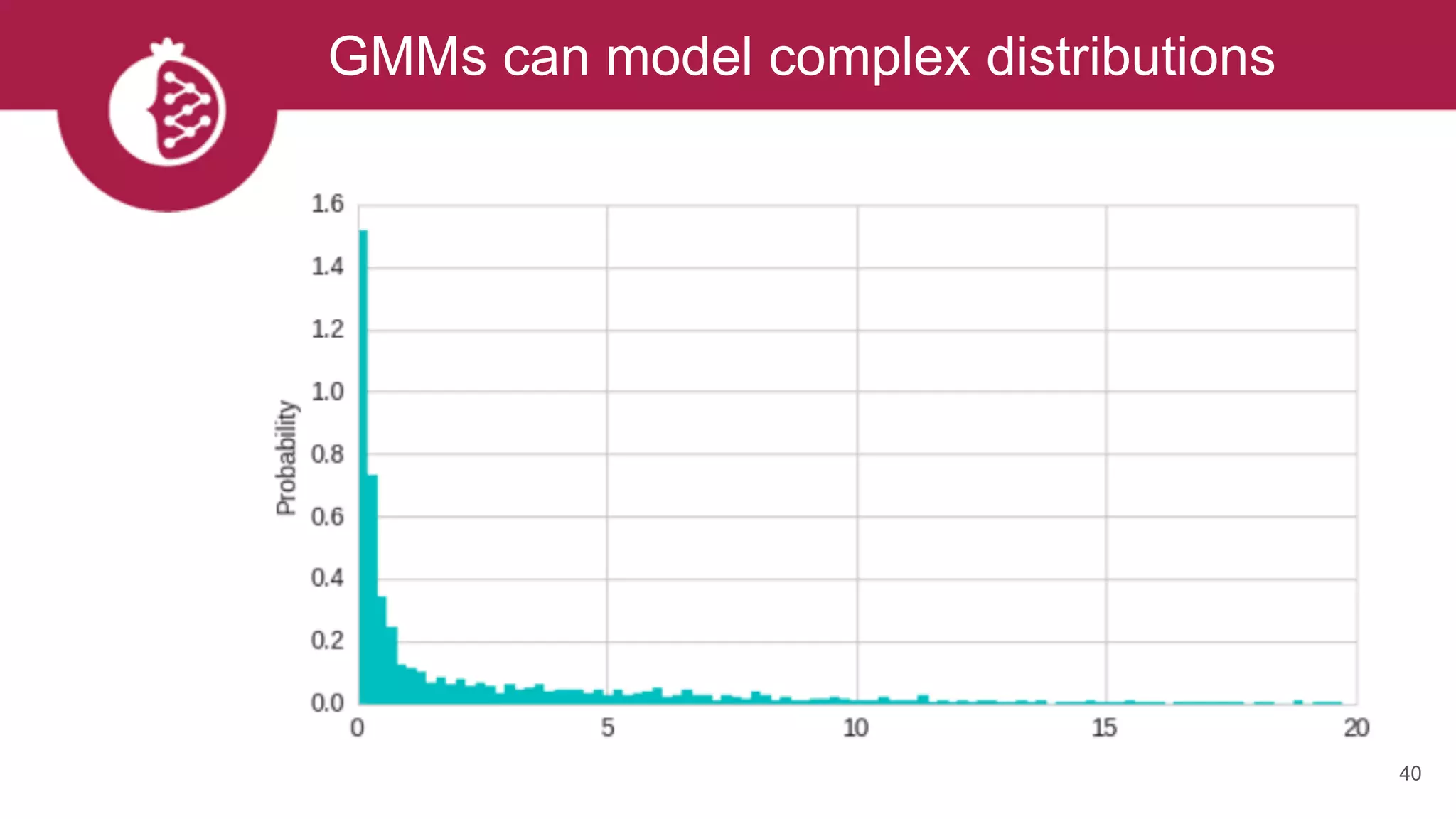
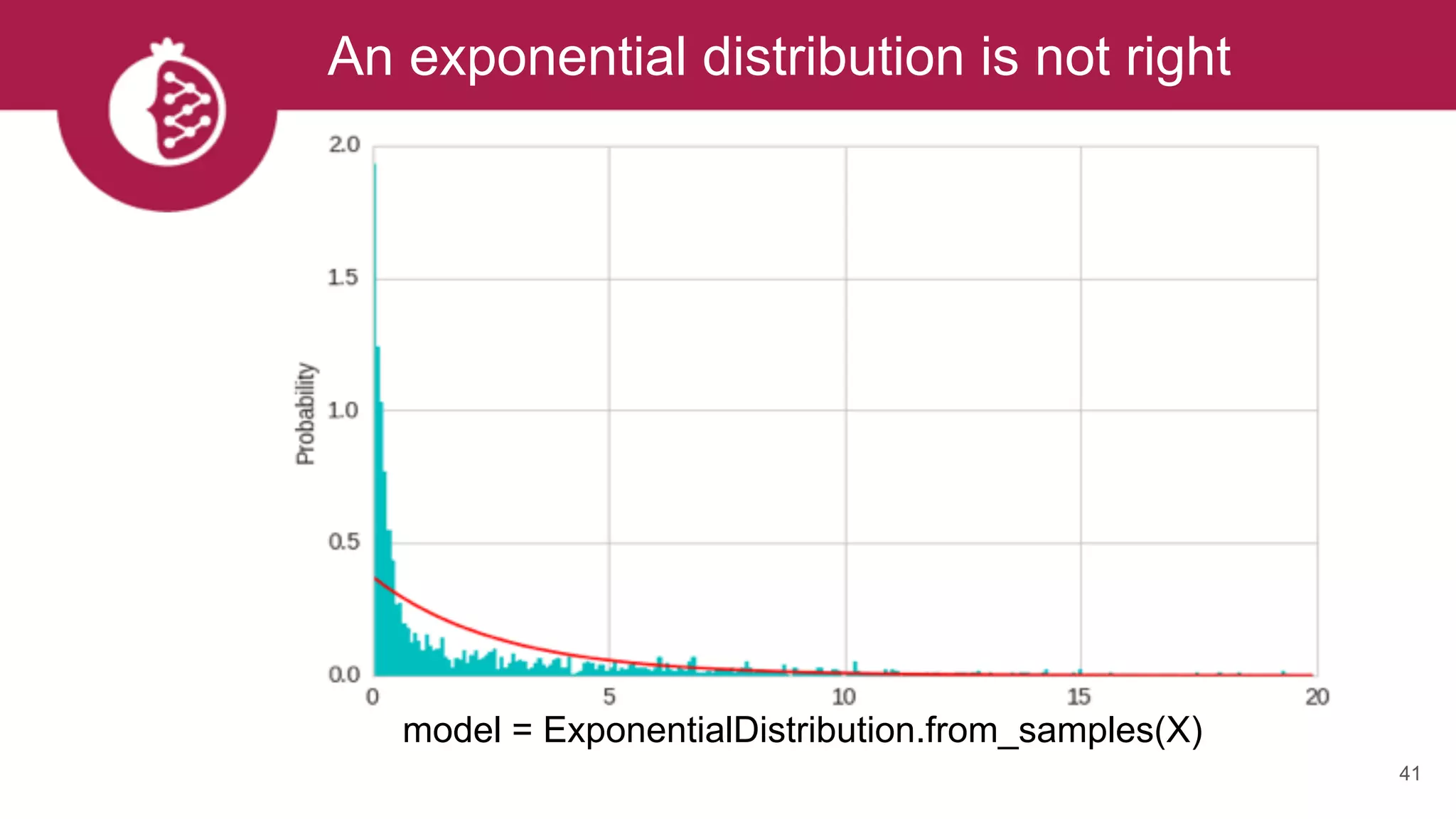
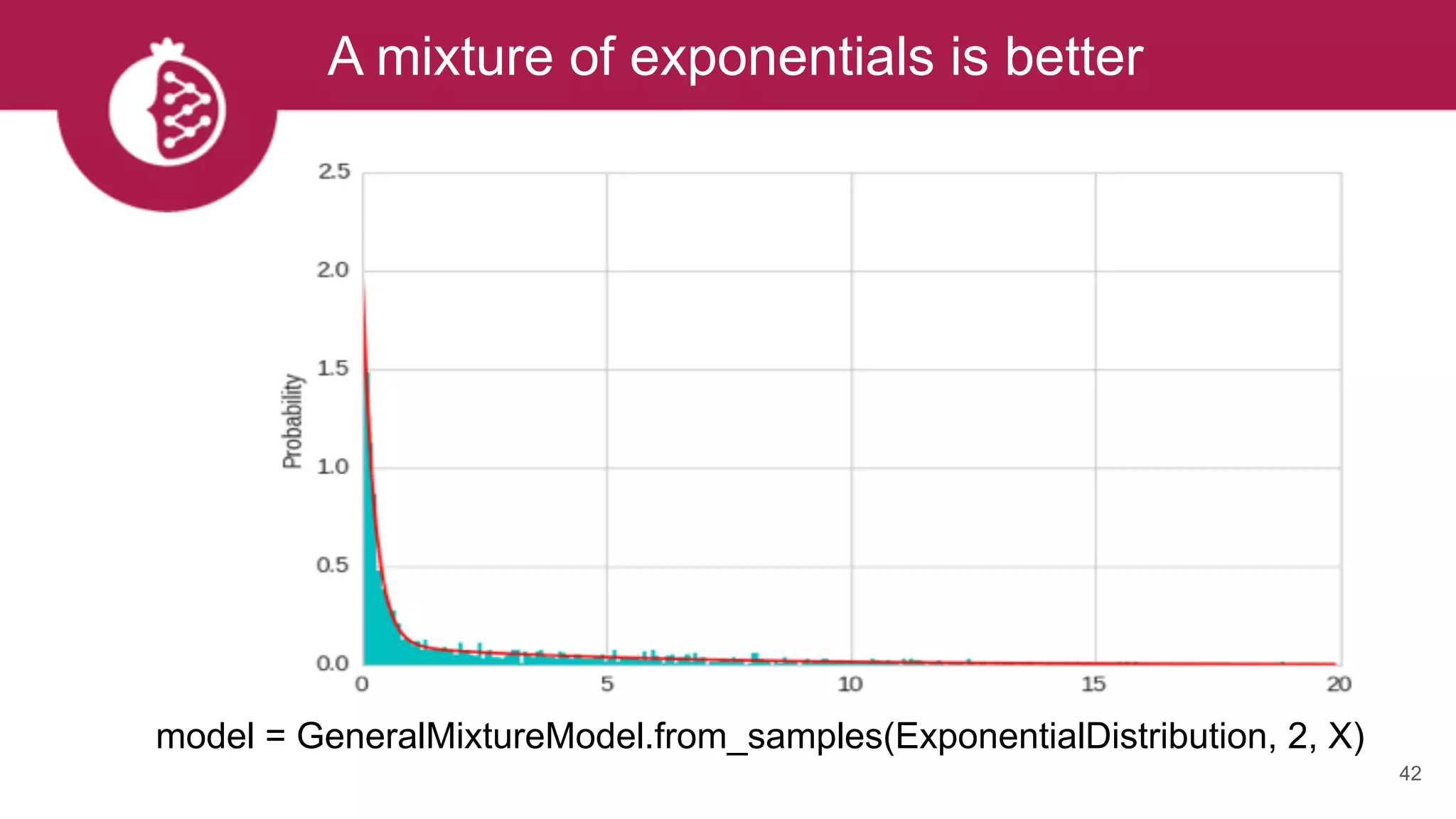
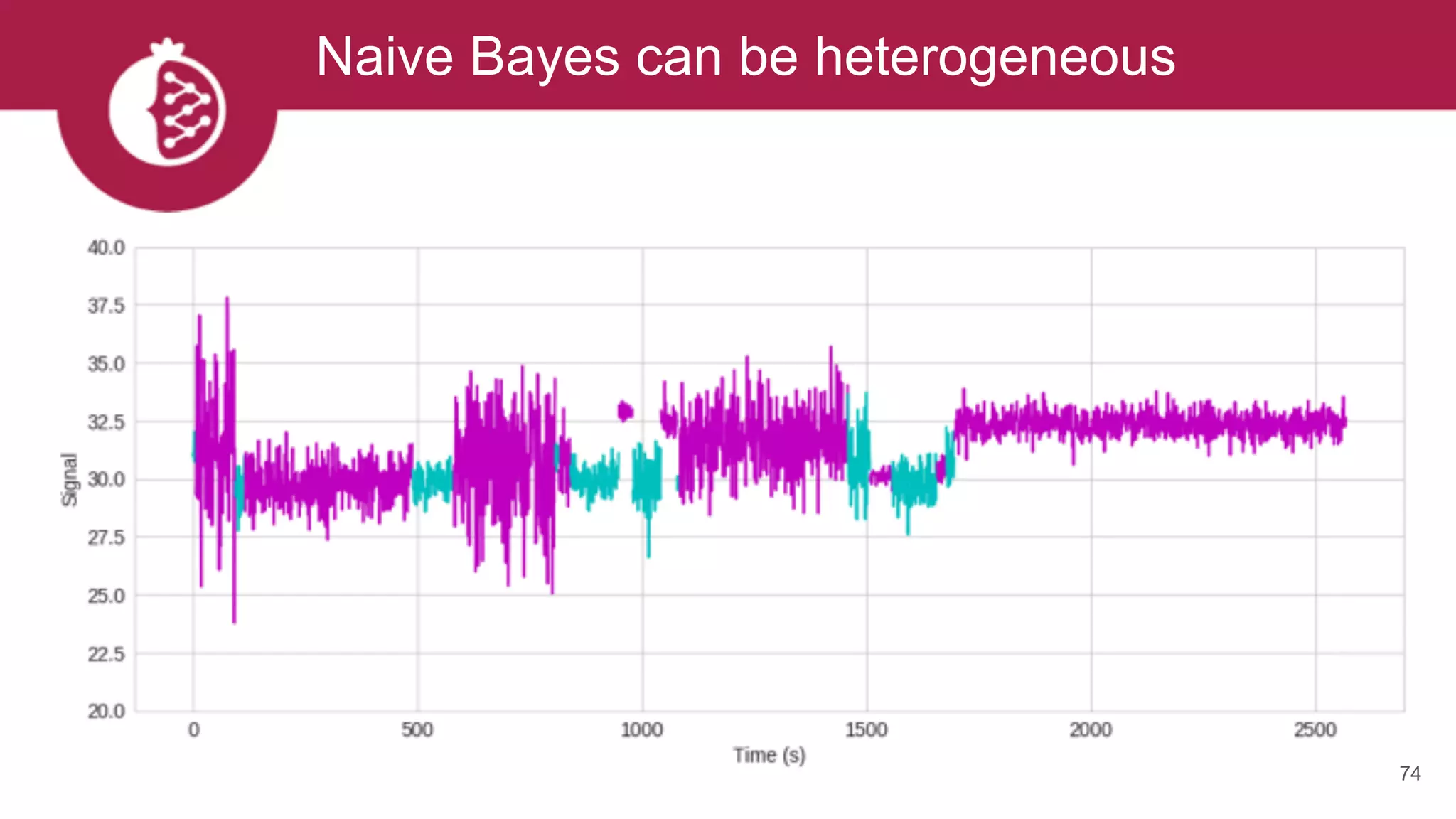
![Heterogeneous mixtures natively supported
43
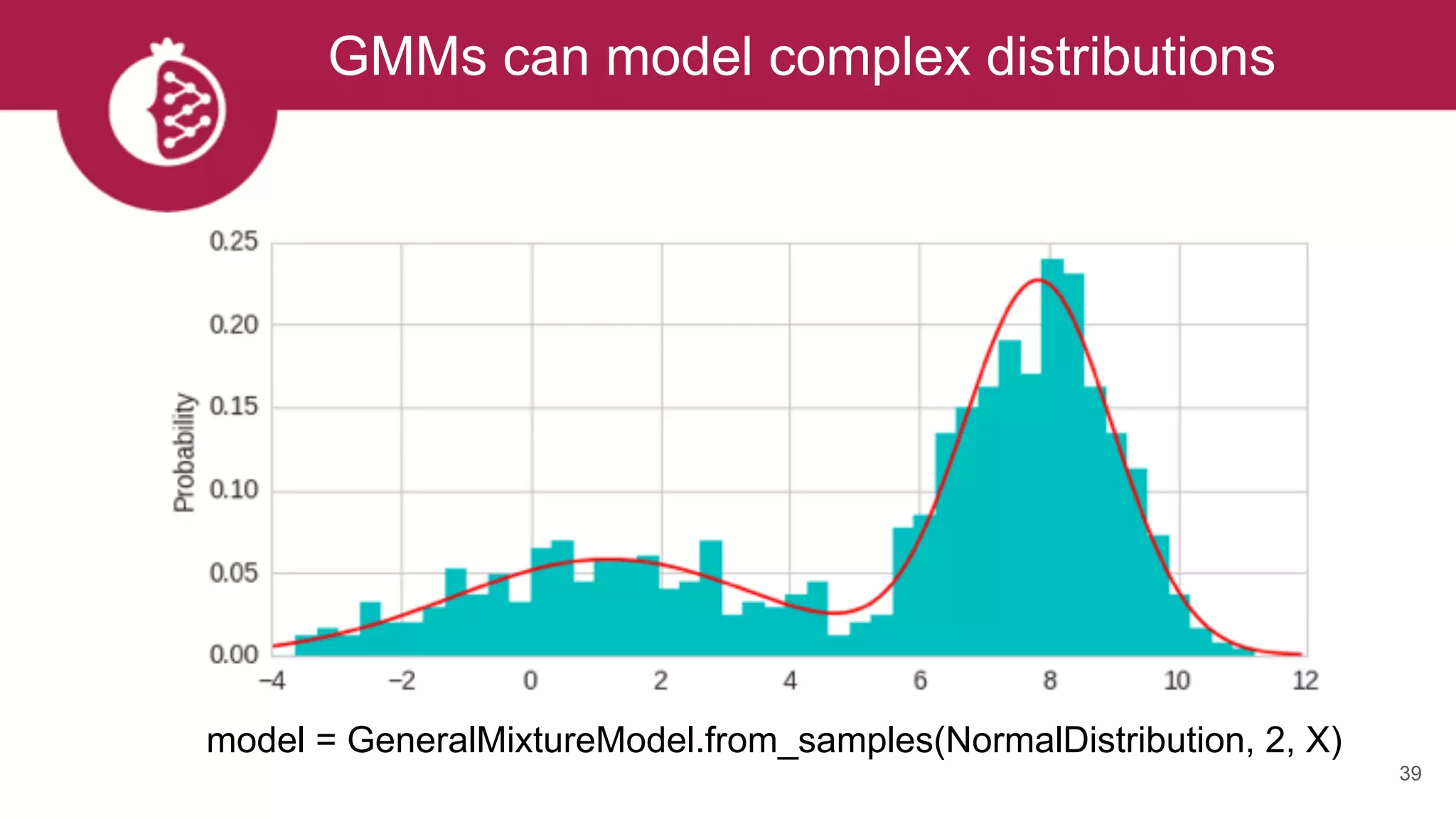
model = GeneralMixtureModel.from_samples([ExponentialDistribution, UniformDistribution], 2, X)](https://image.slidesharecdn.com/tzxq8ddyrpomx9xvfny5-signature-a56a2239ee4119c26e3e070c0ece32f2898146a45b6997205cf22bd9d4505024-poli-170721202738/75/Maxwell-W-Libbrecht-pomegranate-fast-and-flexible-probabilistic-modeling-in-python-43-2048.jpg)
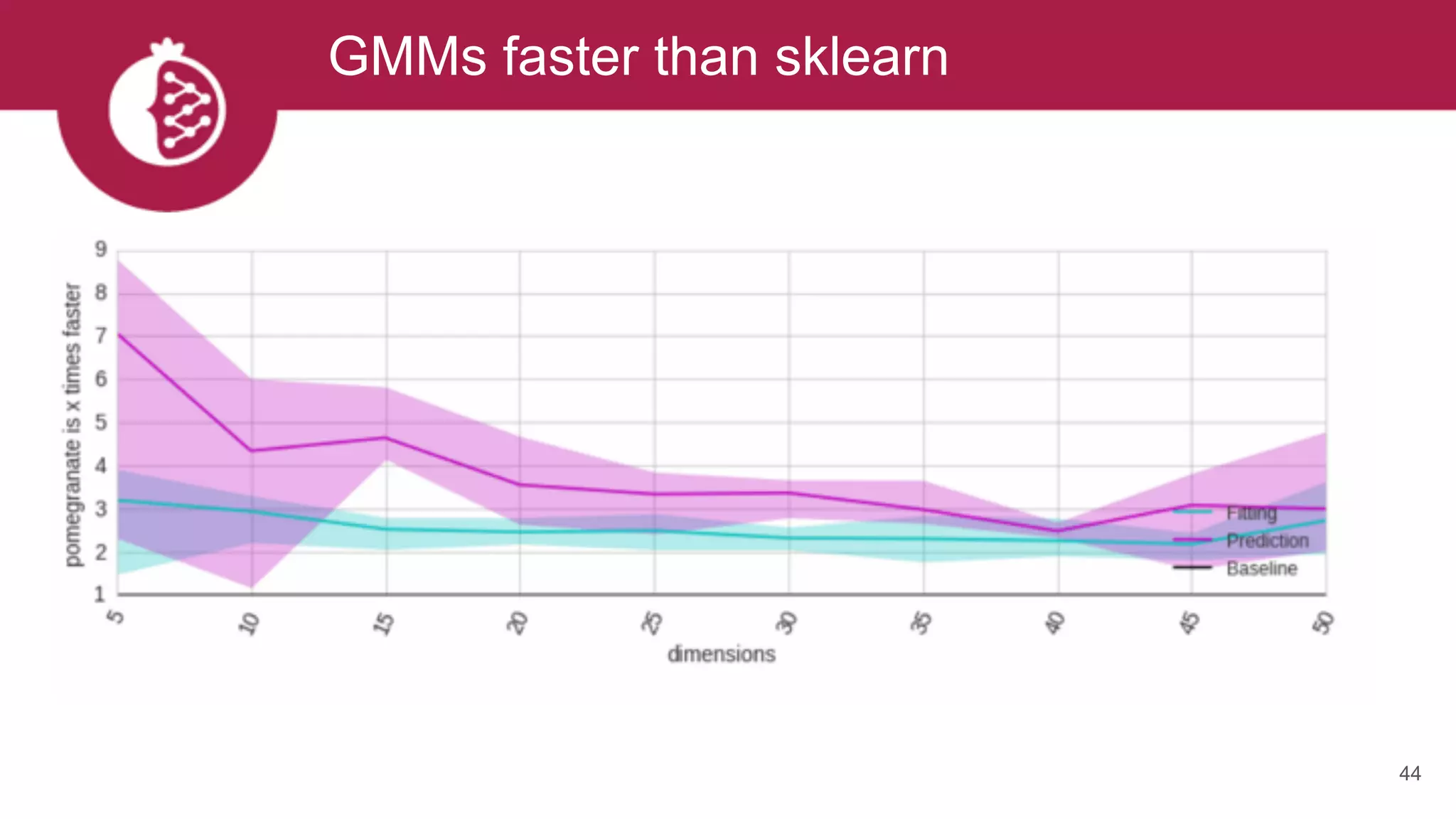





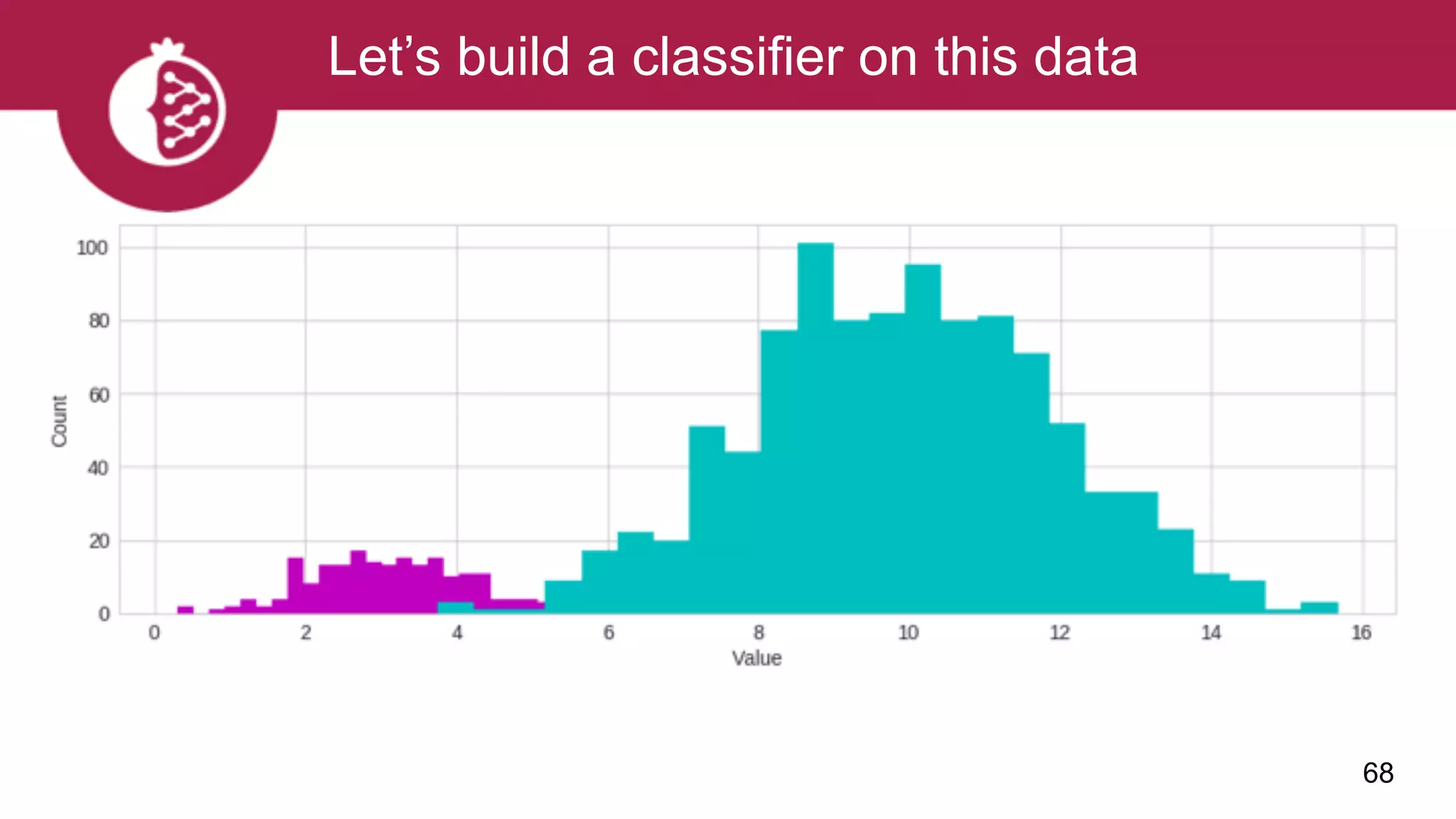
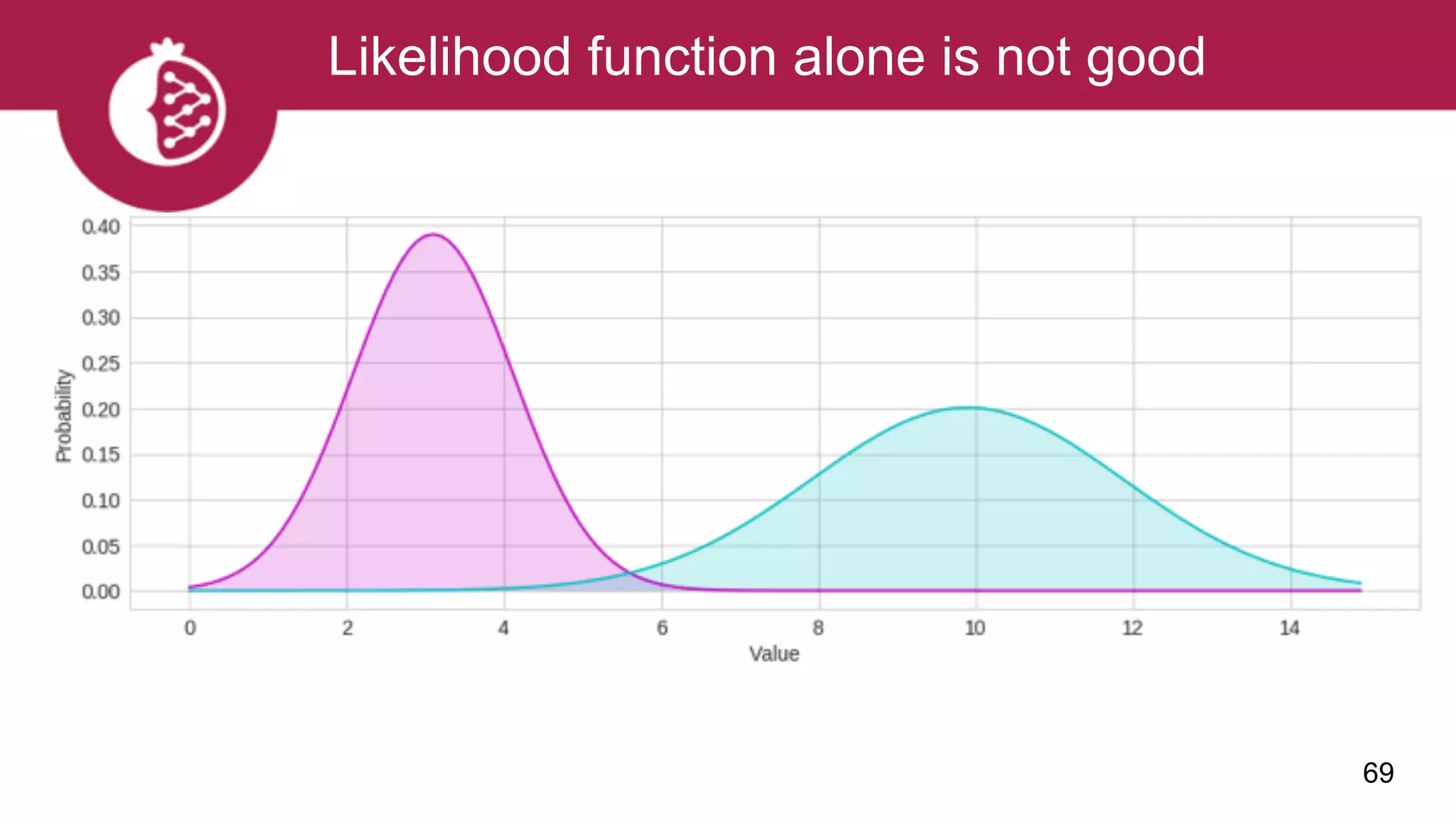
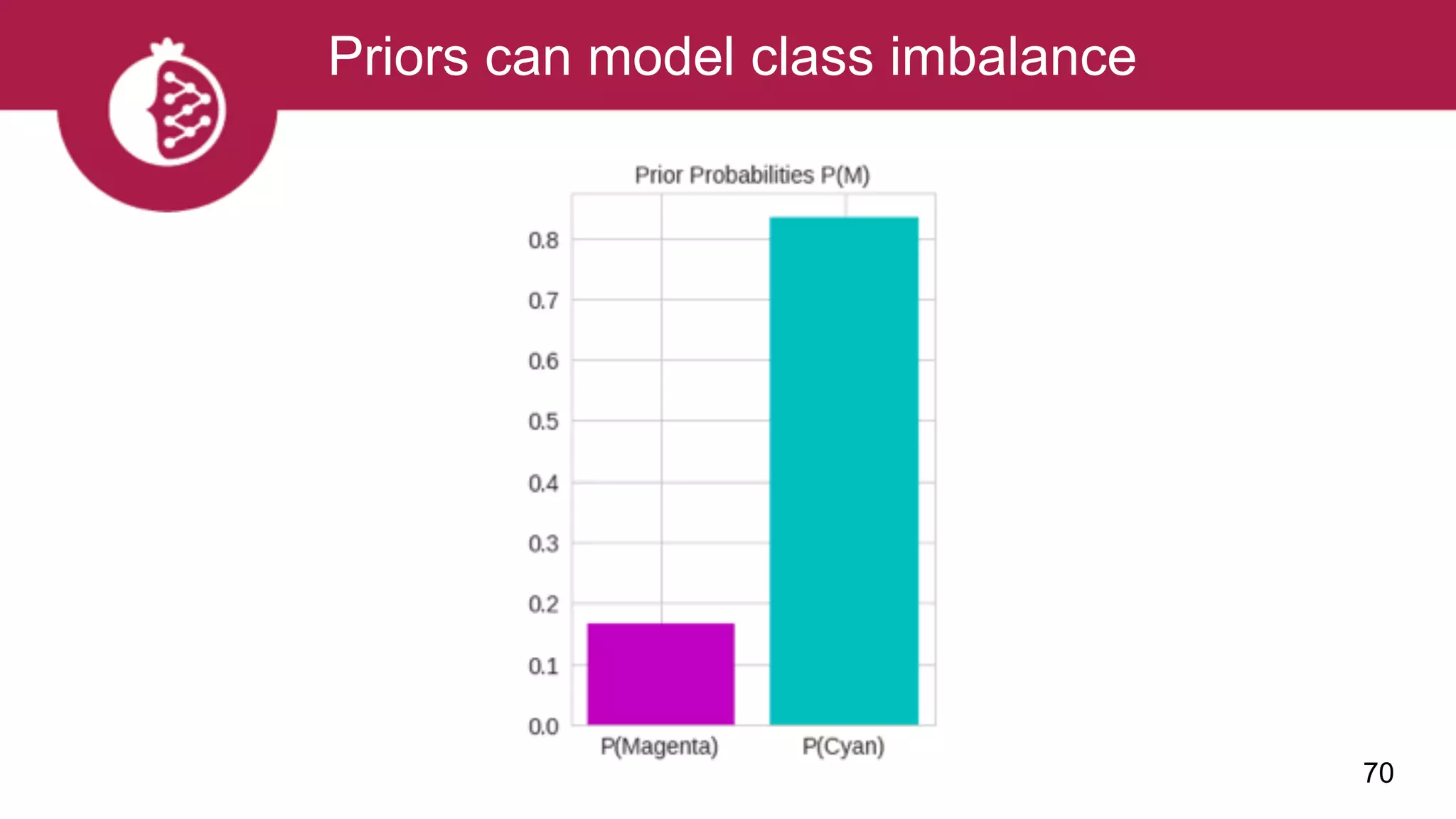
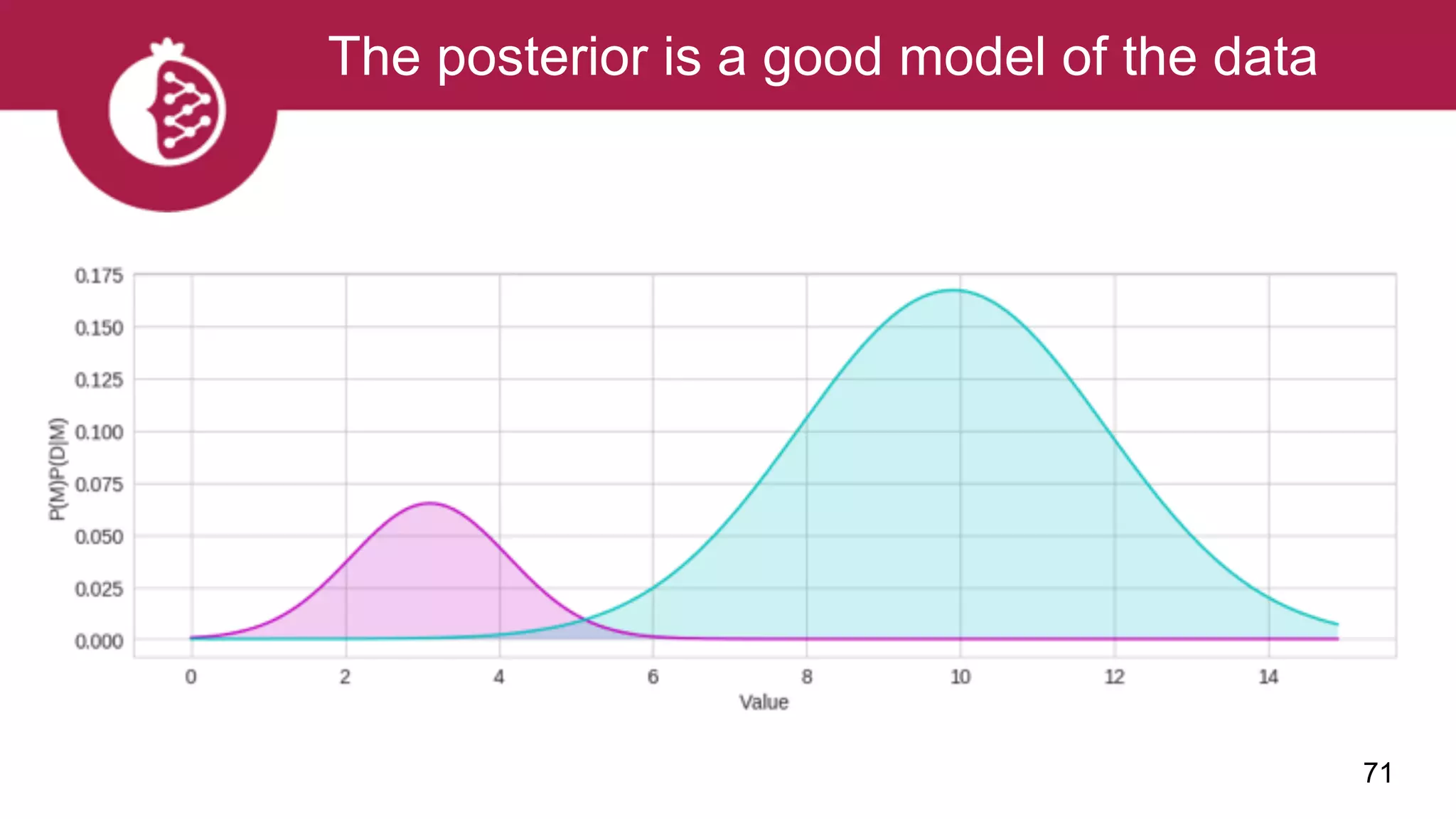
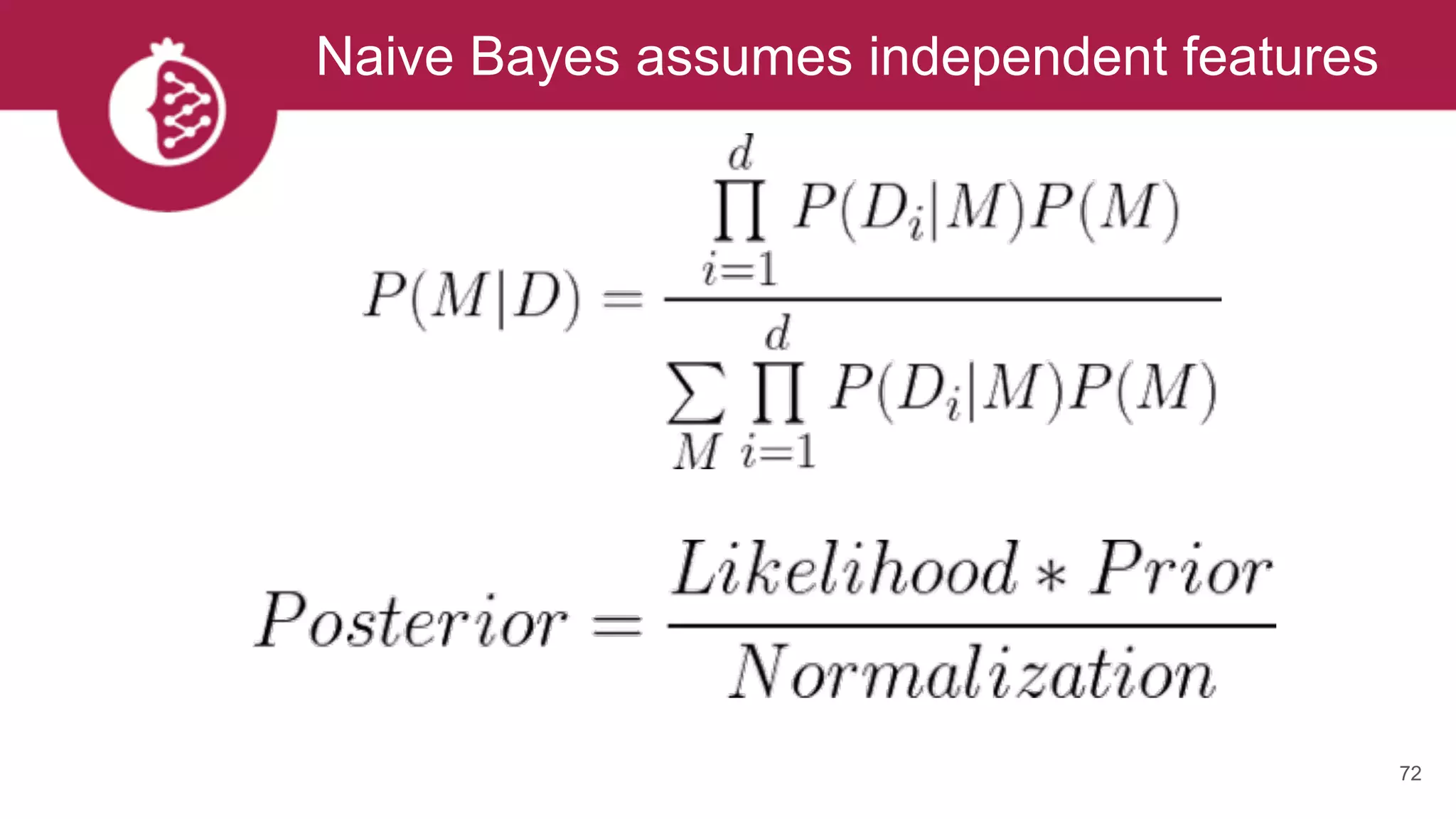
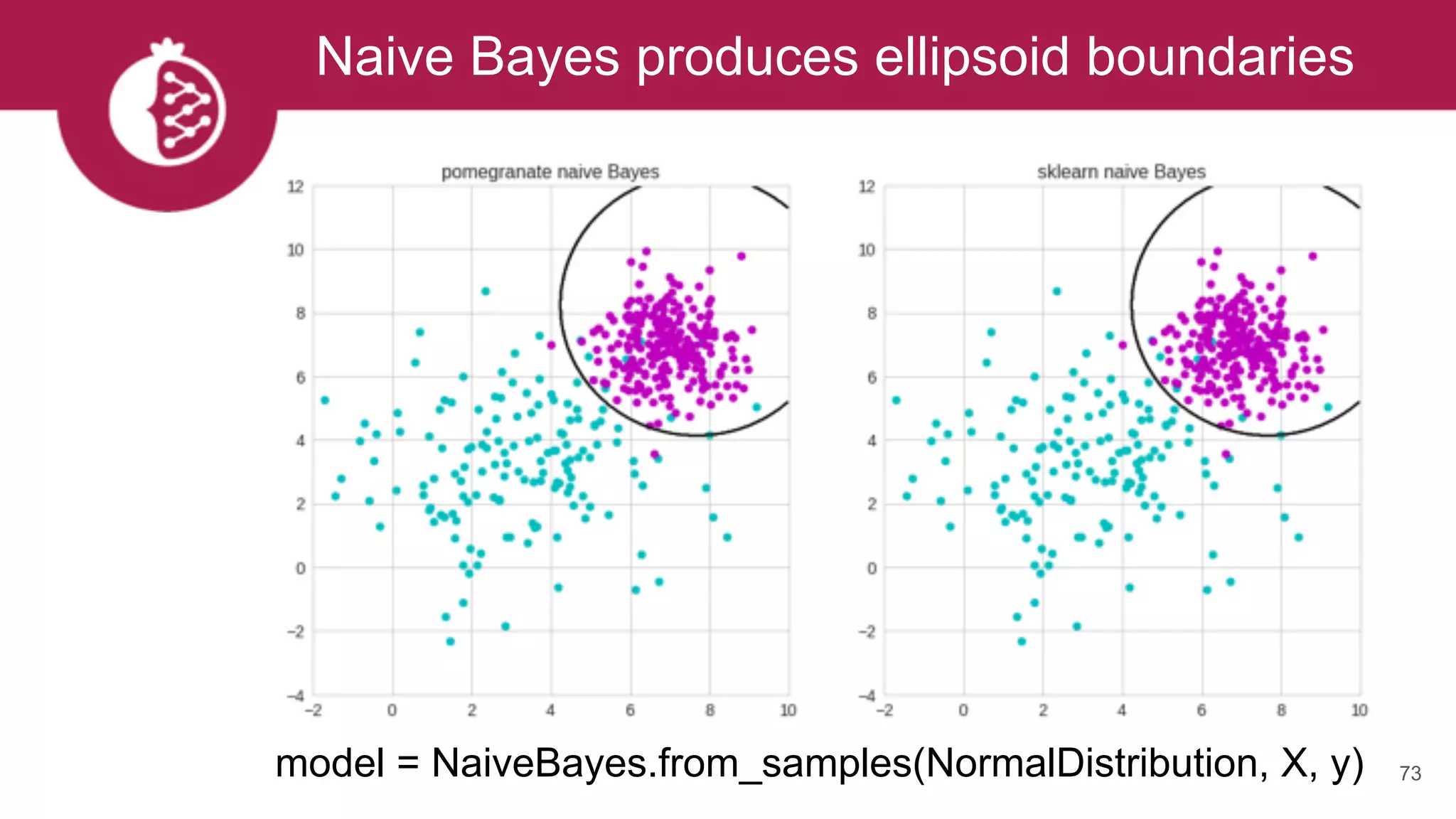
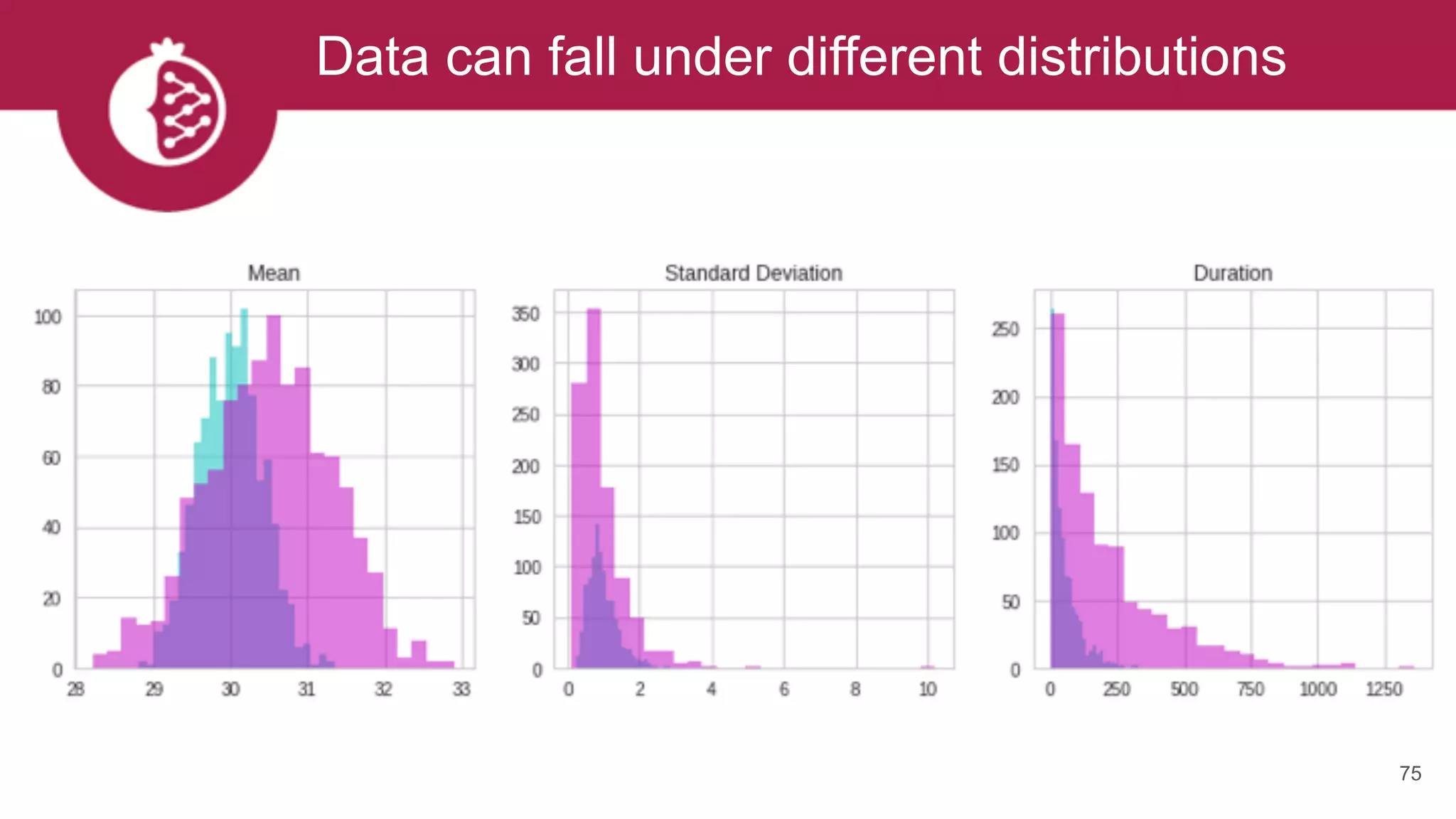
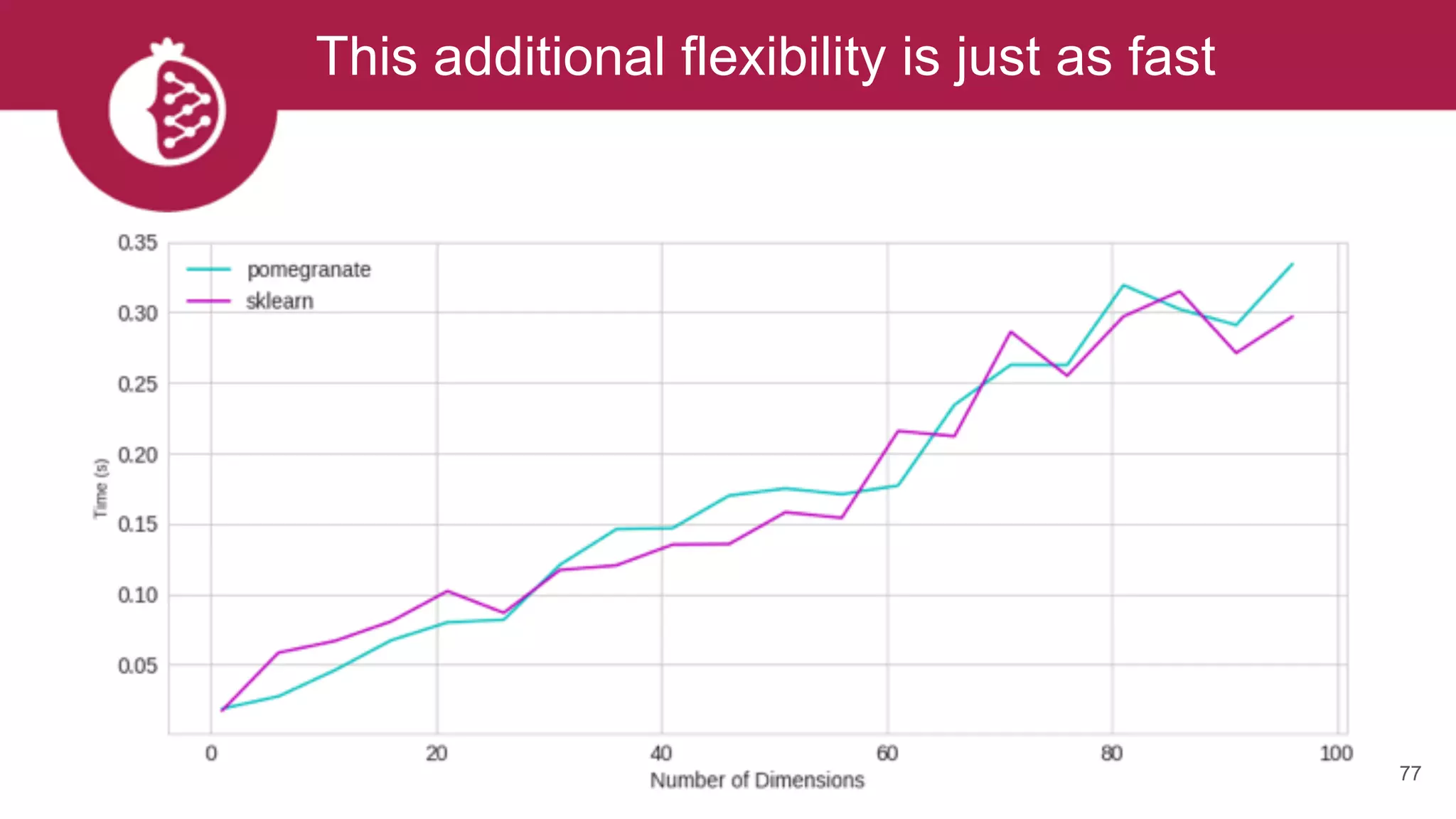
![Using appropriate distributions is better
76
model = NaiveBayes.from_samples(NormalDistribution, X_train, y_train)
print "Gaussian Naive Bayes: ", (model.predict(X_test) == y_test).mean()
clf = GaussianNB().fit(X_train, y_train)
print "sklearn Gaussian Naive Bayes: ", (clf.predict(X_test) == y_test).mean()
model = NaiveBayes.from_samples([NormalDistribution, LogNormalDistribution,
ExponentialDistribution], X_train, y_train
print "Heterogeneous Naive Bayes: ", (model.predict(X_test) == y_test).mean()
Gaussian Naive Bayes: 0.798
sklearn Gaussian Naive Bayes: 0.798
Heterogeneous Naive Bayes: 0.844](https://image.slidesharecdn.com/tzxq8ddyrpomx9xvfny5-signature-a56a2239ee4119c26e3e070c0ece32f2898146a45b6997205cf22bd9d4505024-poli-170721202738/75/Maxwell-W-Libbrecht-pomegranate-fast-and-flexible-probabilistic-modeling-in-python-76-2048.jpg)
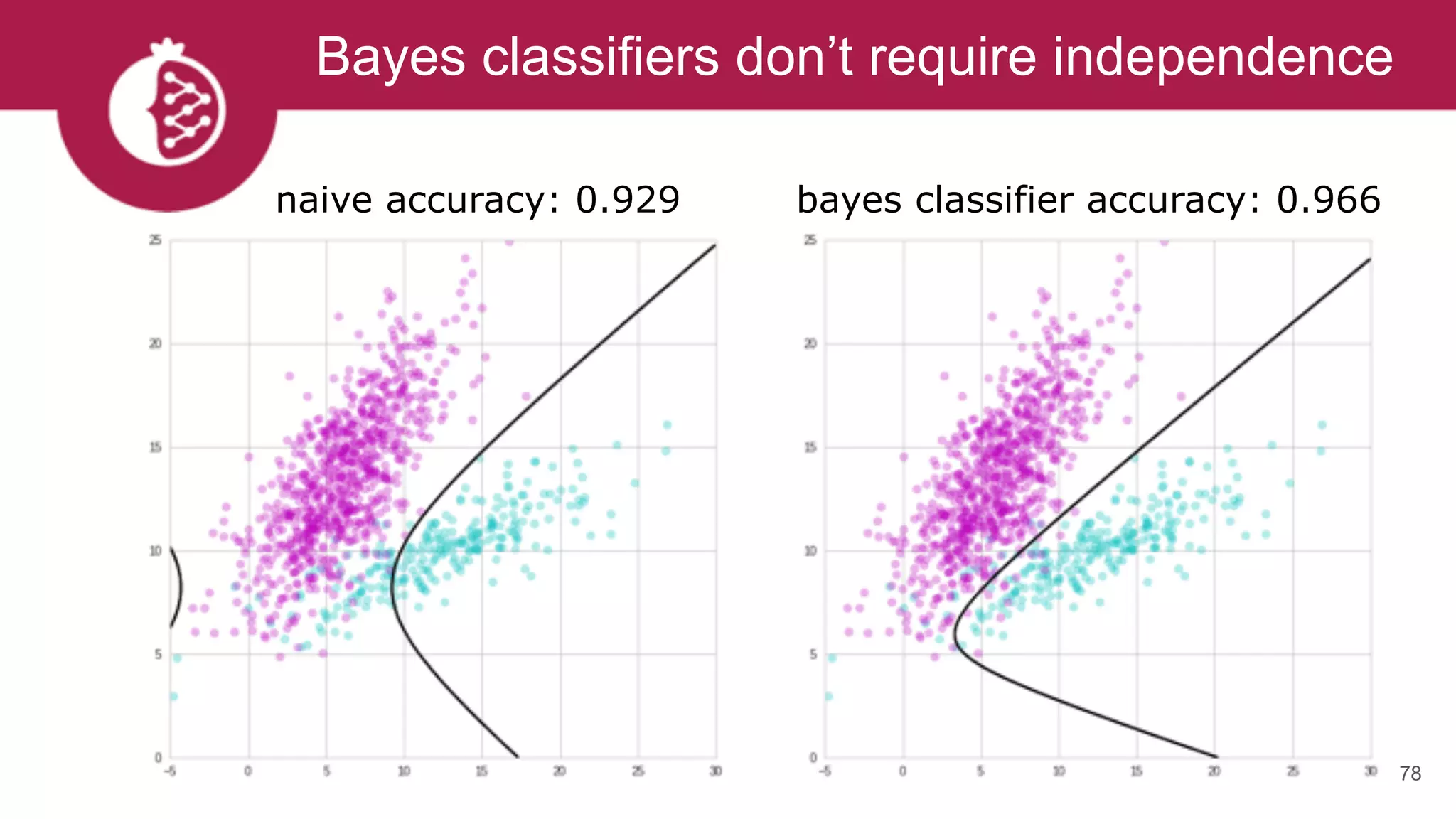
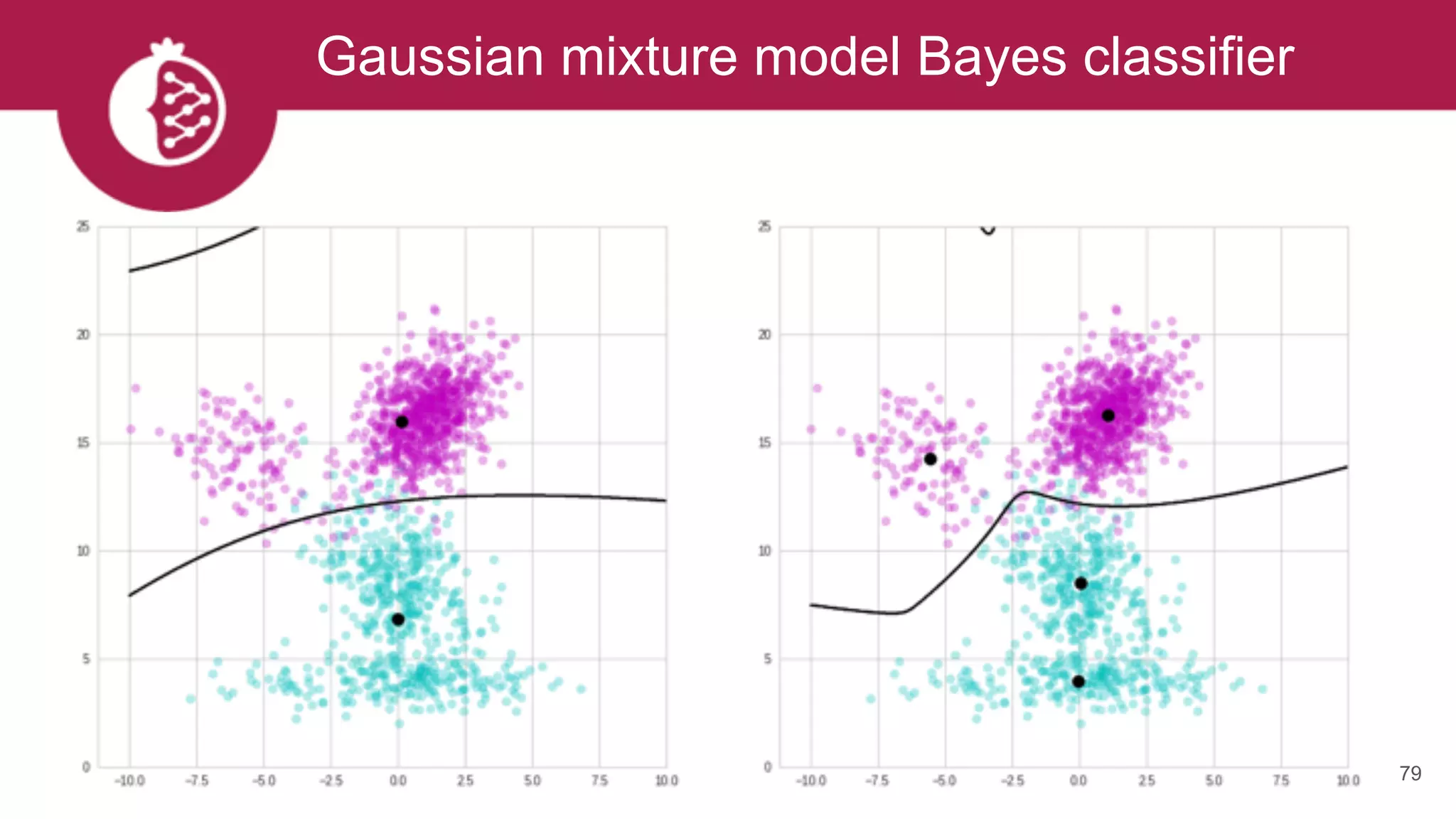
![Creating complex Bayes classifiers is easy
80
gmm_a = GeneralMixtureModel.from_samples(MultivariateGaussianDistribution, 2, X[y == 0])
gmm_b = GeneralMixtureModel.from_samples(MultivariateGaussianDistribution, 2, X[y == 1])
model_b = BayesClassifier([gmm_a, gmm_b], weights=numpy.array([1-y.mean(), y.mean()]))](https://image.slidesharecdn.com/tzxq8ddyrpomx9xvfny5-signature-a56a2239ee4119c26e3e070c0ece32f2898146a45b6997205cf22bd9d4505024-poli-170721202738/75/Maxwell-W-Libbrecht-pomegranate-fast-and-flexible-probabilistic-modeling-in-python-80-2048.jpg)
![Creating complex Bayes classifiers is easy
81
mc_a = MarkovChain.from_samples(X[y == 0])
mc_b = MarkovChain.from_samples(X[y == 1])
model_b = BayesClassifier([mc_a, mc_b], weights=numpy.array([1-y.mean(), y.mean()]))
hmm_a = HiddenMarkovModel.from_samples(X[y == 0])
hmm_b = HiddenMarkovModel.from_samples(X[y == 1])
model_b = BayesClassifier([hmm_a, hmm_b], weights=numpy.array([1-y.mean(), y.mean()]))
bn_a = BayesianNetwork.from_samples(X[y == 0])
bn_b = BayesianNetwork.from_samples(X[y == 1])
model_b = BayesClassifier([bn_a, bn_b], weights=numpy.array([1-y.mean(), y.mean()]))](https://image.slidesharecdn.com/tzxq8ddyrpomx9xvfny5-signature-a56a2239ee4119c26e3e070c0ece32f2898146a45b6997205cf22bd9d4505024-poli-170721202738/75/Maxwell-W-Libbrecht-pomegranate-fast-and-flexible-probabilistic-modeling-in-python-81-2048.jpg)