This document discusses basic loops and functions in R programming. It covers control statements like loops and if/else, arithmetic and boolean operators, default argument values, and returning values from functions. It also describes R programming structures, recursion, and provides an example of implementing quicksort recursively and constructing a binary search tree. The key topics are loops, control flow, functions, recursion, and examples of sorting and binary trees.



![Control Statements
Loops
Basic Structure
In our function oddcount() the line
for (n in x) {
will be instantly recognized by Python programmers. It of course means that
there will be one iteration of
the loop for each component of the vector x, with n taking on the values of those
components.
In otherwords, in the first iteration, n = x[1], in the second iteration n = x[2], etc.
For example:
> x <- c(5,12,13)
> for (n in x) print(nˆ2)
[1] 25
[1] 144
[1] 169
C-style looping with while and repeat are also available, complete with break:](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-4-320.jpg)
![> i <- 1
> while(1) {
+ i <- i+4
+ if (i > 10) break
+ }
> i
[1] 13
Of course, break can be used with for too.
Another useful statement is next, which instructs the interpreter to go to the next
iteration of the loop.
Usage of this construct often allows one to avoid using complexly nested if-then-
else statements, which make the
code confusing.](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-5-320.jpg)
![Looping Over Nonvector Sets
The for construct works on any vector, regardless of mode. One can loop over a
vector of file names, for
instance. Say we have files x and y with contents
1
2
3
4
5
6
and
5
12
13
Then this loop prints each of them:
> for (fn in c("x","y")) print(scan(fn))
Read 6 items
[1] 1 2 3 4 5 6
Read 3 items
[1] 5 12 13
R does not directly support iteration over nonvector sets, but there are indirect
yet easy ways to accomplish
it. One way would be to use lapply()](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-6-320.jpg)
![get(), as in the
following example. Here we have two matrices, u and v, containing statistical data,
and we wish to apply R
linear regression function lm() to each of them:
> u
[,1] [,2]
[1,] 1 1
[2,] 2 2
[3,] 3 4
> v
[,1] [,2]
[1,] 8 15
[2,] 12 10
[3,] 20 2
> for (m in c("u","v")) {
+ z <- get(m)
+ print(lm(z[,2] ˜ z[,1]))](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-7-320.jpg)
![Call:
lm(formula = z[, 2] ˜ z[, 1])
Coefficients:
(Intercept) z[, 1]
-0.6667 1.5000
Call:
lm(formula = z[, 2] ˜ z[, 1])
Coefficients:
(Intercept) z[, 1]
23.286 -1.071
The reader is welcome to make his/her own refinements here.](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-8-320.jpg)

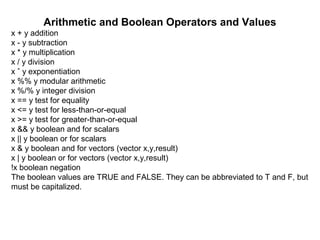
![These values change to 1 and 0 in arithmetic expressions, e.g.
> 1 < 2
[1] TRUE
> (1 < 2) * (3 < 4)
[1] 1
> (1 < 2) * (3 < 4) * (5 < 1)
[1] 0
> (1 < 2) == TRUE
[1] TRUE
> (1 < 2) == 1
[1] TRUE](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-11-320.jpg)
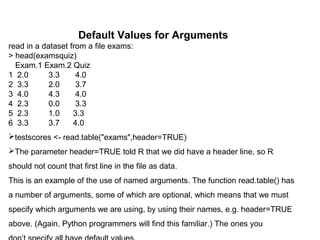





![No Pointers in R
R does not have variables corresponding to pointers or references like those of,
say, the C language. This can make programming more difficult in some cases.
(As of this writing, the current version of R has an experimental feature called
reference classes, which may reduce the difficulty.)
For example, you cannot write a function that directly changes its arguments.
>>> x = [13,5,12]
>>> x.sort()
>>> x
[5, 12, 13]
Here, the value of x, the argument to sort(), changed. By contrast, here’s
how it works in R:
> x <- c(13,5,12)
> sort(x)
[1] 5 12 13
> x
[1] 13 5 12](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-18-320.jpg)


![qs <- function(x) {
if (length(x) <= 1) return(x)
pivot <- x[1]
therest <- x[-1]
sv1 <- therest[therest < pivot]
sv2 <- therest[therest >= pivot]
sv1 <- qs(sv1)
sv2 <- qs(sv2)
return(c(sv1,pivot,sv2))
}
if (length(x) <= 1) return(x)](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-21-320.jpg)

![Extended Example: A Binary Search Tree
stored 8 in the root—that is, the head—of the tree. Its two child nodes contain 5
and 20, and the former itself has two child nodes, which store 2 and 6.
> x <- newtree(8,3)
> x
$mat
[,1] [,2] [,3]
[1,] NA NA 8
[2,] NA NA NA
[3,] NA NA NA
$nxt
[1] 2
$inc
[1] 3
> x <- ins(1,x,5)
> x](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-23-320.jpg)
![$mat
[,1] [,2] [,3]
[1,] 2 NA 8
[2,] NA NA 5
[3,] NA NA NA
$nxt
[1] 3
$inc
[1] 3
> x <- ins(1,x,6)
> x
$mat
[,1] [,2] [,3]
[1,] 2 NA 8
[2,] NA 3 5
[3,] NA NA 6
$nxt
[1] 4
$inc
[1] 3
> x <- ins(1,x,2)
> x](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-24-320.jpg)
![$mat
[,1] [,2] [,3]
[1,] 2 NA 8
[2,] 4 3 5
[3,] NA NA 6
[4,] NA NA 2
[5,] NA NA NA
[6,] NA NA NA
$nxt
[1] 5
$inc
[1] 3
> x <- ins(1,x,20)
> x
$mat
[,1] [,2] [,3]
[1,] 2 5 8
[2,] 4 3 5
[3,] NA NA 6
[4,] NA NA 2
[5,] NA NA 20
[6,] NA NA NA](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-25-320.jpg)
![$nxt
[1] 6
$inc
[1] 3
What happened here? First, the command containing our call newtree(8,3) creates
a new tree, assigned to x, storing the number 8. The argument 3 specifies that we
allocate storage room three rows at a time.
The result is that the matrix component of the list x is now as follows:
[,1] [,2] [,3]
[1,] NA NA 8
[2,] NA NA NA
[3,] NA NA NA](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-26-320.jpg)

![NOTE This example is for the purpose of demonstrating recursion. R’s own sort
function,
sort(), is much faster, as it is written in C.
qs <- function(x) {
if (length(x) <= 1) return(x)
pivot <- x[1]
therest <- x[-1]
sv1 <- therest[therest < pivot]
sv2 <- therest[therest >= pivot]
sv1 <- qs(sv1)
sv2 <- qs(sv2)
return(c(sv1,pivot,sv2))
}
if (length(x) <= 1) return(x)
Without this, the function would keep calling itself repeatedly on empty vectors,
executing forever.](https://image.slidesharecdn.com/loopsandfunctionsinr-181101062724/85/Loops-and-functions-in-r-28-320.jpg)
