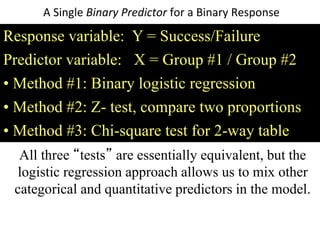
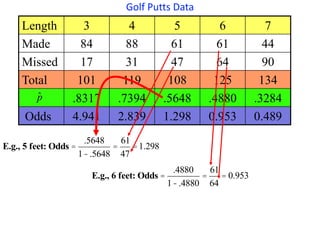
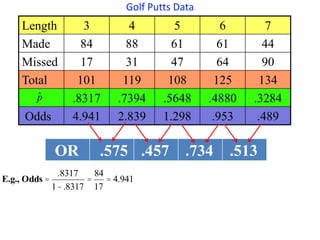
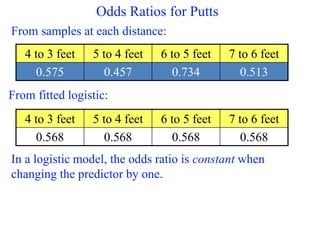
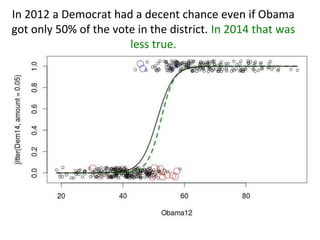
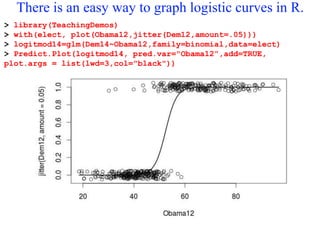
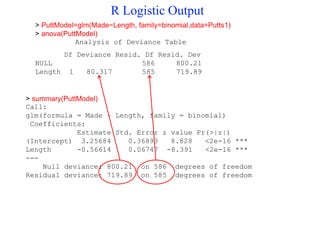
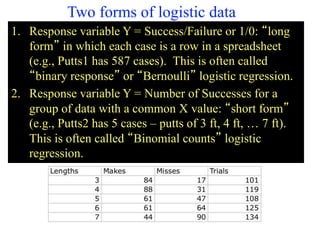
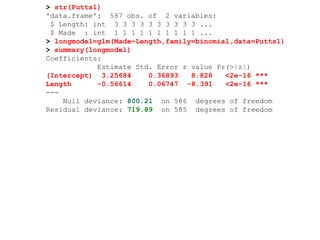
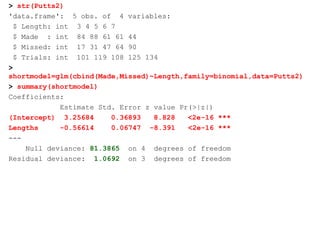
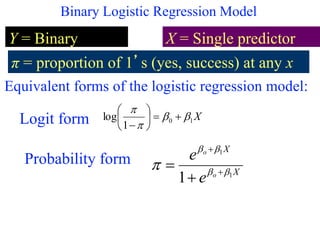
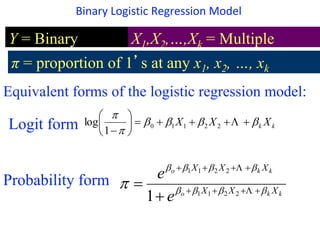
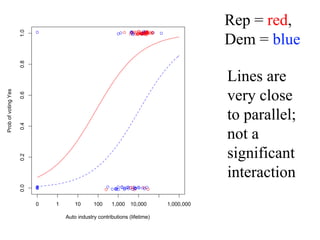
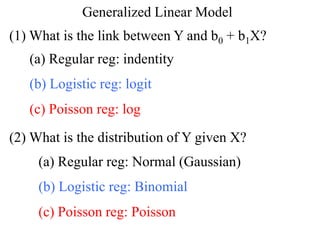
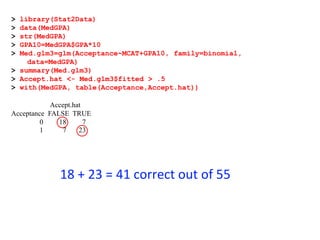
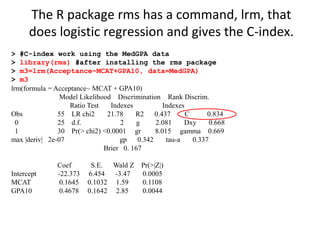
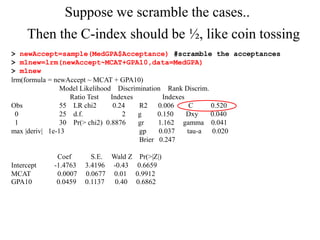
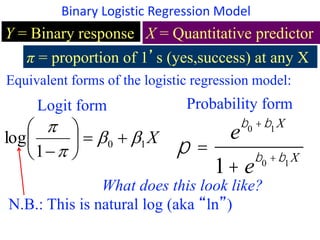
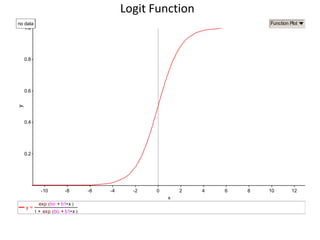
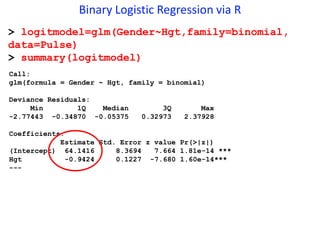
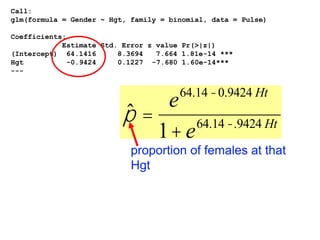
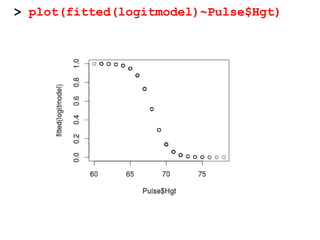
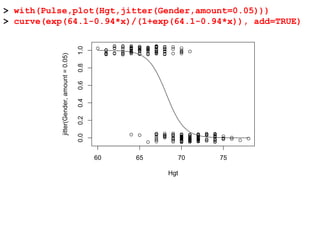
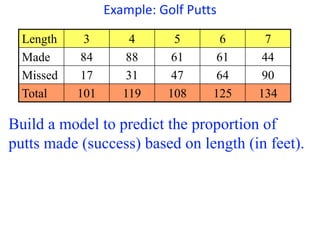
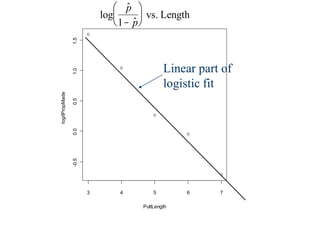
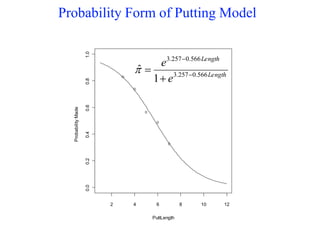
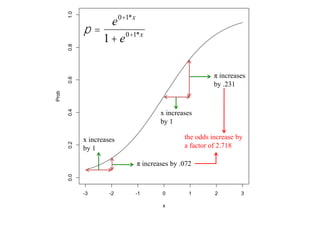
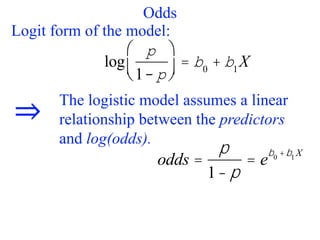
This document provides an overview of logistic regression for categorical response variables. It begins with examples of binary, ordinal, and nominal categorical variables. It then demonstrates how logistic regression models the log-odds of an event rather than the mean, as linear regression does. The key aspects of binary logistic regression models are described, including the logit form and probability form of the model. Examples are provided using R to model gender based on height, the success of golf putts based on length, and the effectiveness of a medical treatment compared to placebo. Key outputs from logistic regression in R like coefficients, odds ratios, and model comparison are also summarized.




![Fair die
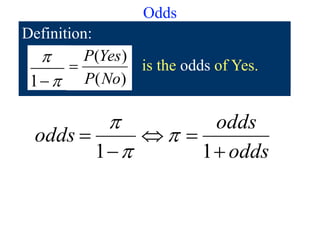
Prob OddsEvent
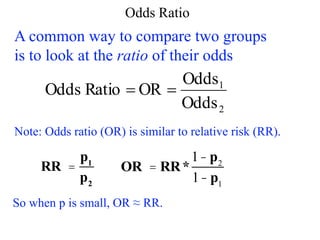
roll a 2 1/6 1/5 [or 1/5:1 or 1:5]
even # 1/2 1 [or 1:1]
X > 2 2/3 2 [or 2:1]](https://image.slidesharecdn.com/logisticregression-200831141028/85/Logisticregression-18-320.jpg)

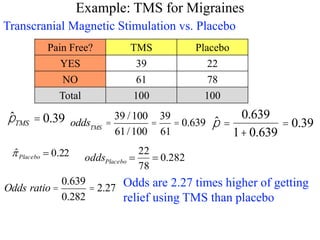
![> datatable=rbind(c(39,22),c(61,78))
> datatable
[,1] [,2]
[1,] 39 22
[2,] 61 78
> chisq.test(datatable,correct=FALSE)
Pearson's Chi-squared test
data: datatable
X-squared = 6.8168, df = 1, p-value = 0.00903
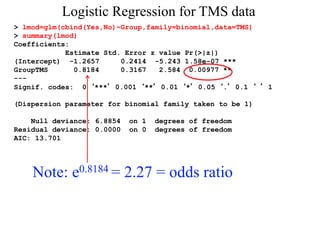
> lmod=glm(cbind(Yes,No)~Group,family=binomial,data=TMS)
> summary(lmod)
Call:
glm(formula = cbind(Yes, No) ~ Group, family = binomial)Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.2657 0.2414 -5.243 1.58e-07 ***
GroupTMS 0.8184 0.3167 2.584 0.00977 **
Binary Logistic Regression
Chi-Square Test for
2-way table](https://image.slidesharecdn.com/logisticregression-200831141028/85/Logisticregression-25-320.jpg)