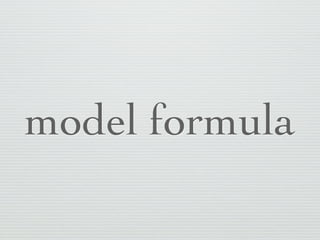
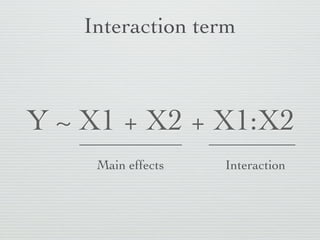
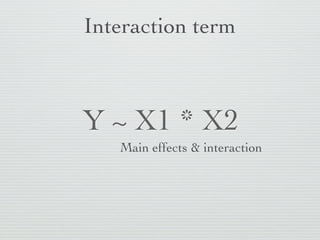
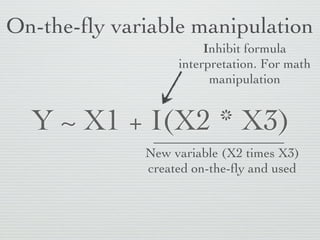
1: This document discusses linear regression analysis in R, including preparing data, specifying models, and interpreting results.
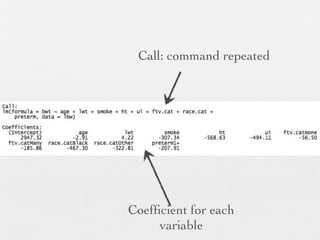
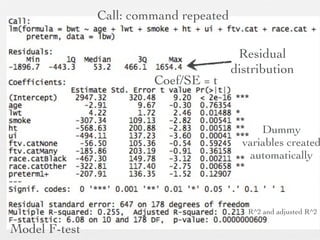
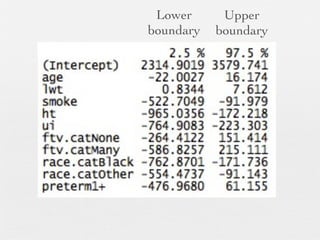
2: Key steps covered include recoding and transforming variables, specifying linear regression models using formula syntax, fitting models using lm(), and interpreting model outputs like coefficients, confidence intervals, and ANOVA tables.
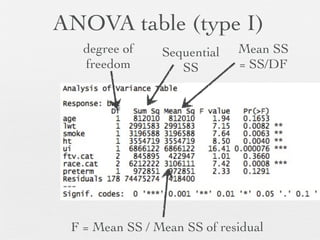
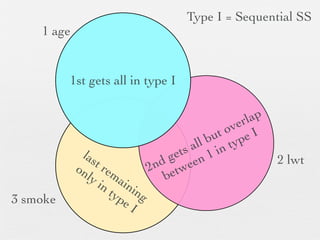
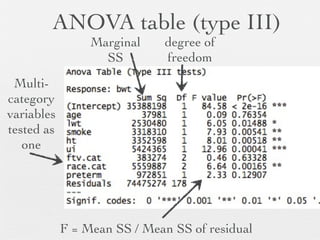
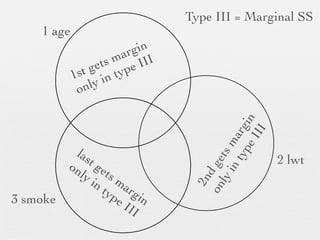
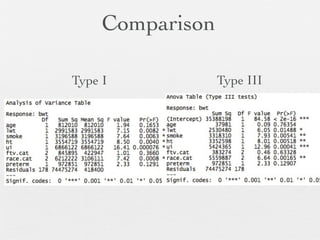
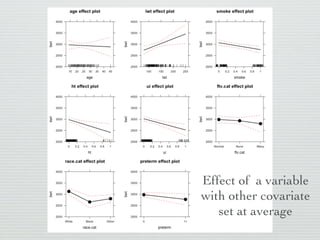
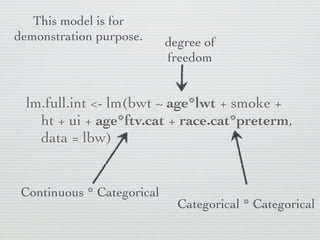
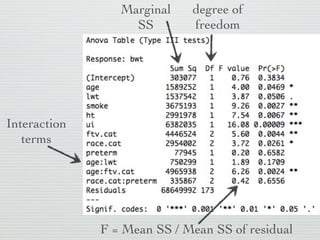
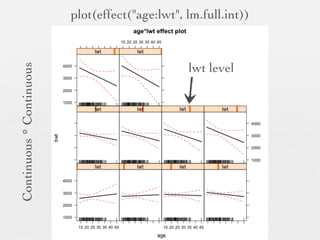
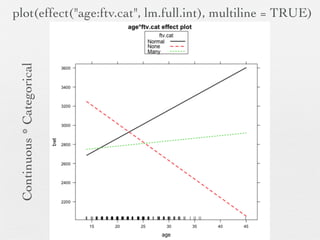
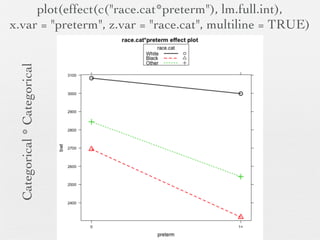
3: Interactions are discussed, along with visualizing effects using the effects package. Both type I and type III ANOVA tables are demonstrated along with distinguishing their interpretations.







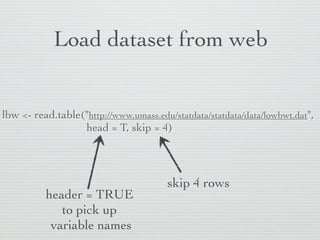
![“Fix” dataset
lbw[c(10,39), "BWT"] <- c(2655, 3035)
BWT column
Replace data points
10th,39th to make the dataset identical
rows to BIO213 dataset](https://image.slidesharecdn.com/20121207linearregression-121202222916-phpapp02/85/Linear-regression-with-R-1-11-320.jpg)
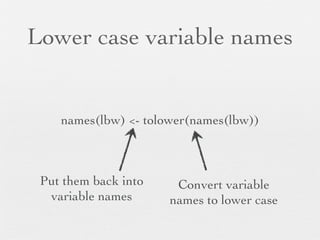







![Numeric to categorical:
range to element
lbw <- within(lbw, {
1st will be reference
## Relabel race
race.cat <- factor(race, levels = 1:3, labels = c("White","Black","Other"))
## Categorize ftv (frequency of visit)
ftv.cat <- cut(ftv, breaks = c(-Inf, 0, 2, Inf), labels = c("None","Normal","Many"))
ftv.cat <- relevel(ftv.cat, ref = "Normal")
## Dichotomize ptl
preterm <- factor(ptl >= 1, levels = c(F,T), labels = c("0","1+"))
}) How breaks work
(-Inf 0] 1 2] 3 4 5 6 Inf ]
None Normal Many](https://image.slidesharecdn.com/20121207linearregression-121202222916-phpapp02/85/Linear-regression-with-R-1-21-320.jpg)


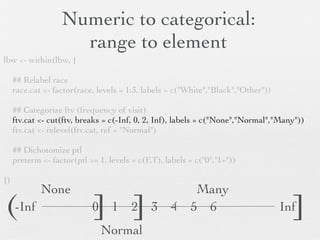
![Binary 0,1 to No,Yes
lbw <- within(lbw, {
## Categorize smoke ht ui
smoke <- factor(smoke, levels = 0:1, labels = c("No","Yes")) One-by-one
ht <- factor(ht, levels = 0:1, labels = c("No","Yes"))
ui <- factor(ui, levels = 0:1, labels = c("No","Yes")) method
})
## Alternative to above
lbw[,c("smoke","ht","ui")] <-
lapply(lbw[,c("smoke","ht","ui")],
function(var) { Loop method
var <- factor(var, levels = 0:1, labels = c("No","Yes"))
})](https://image.slidesharecdn.com/20121207linearregression-121202222916-phpapp02/85/Linear-regression-with-R-1-24-320.jpg)