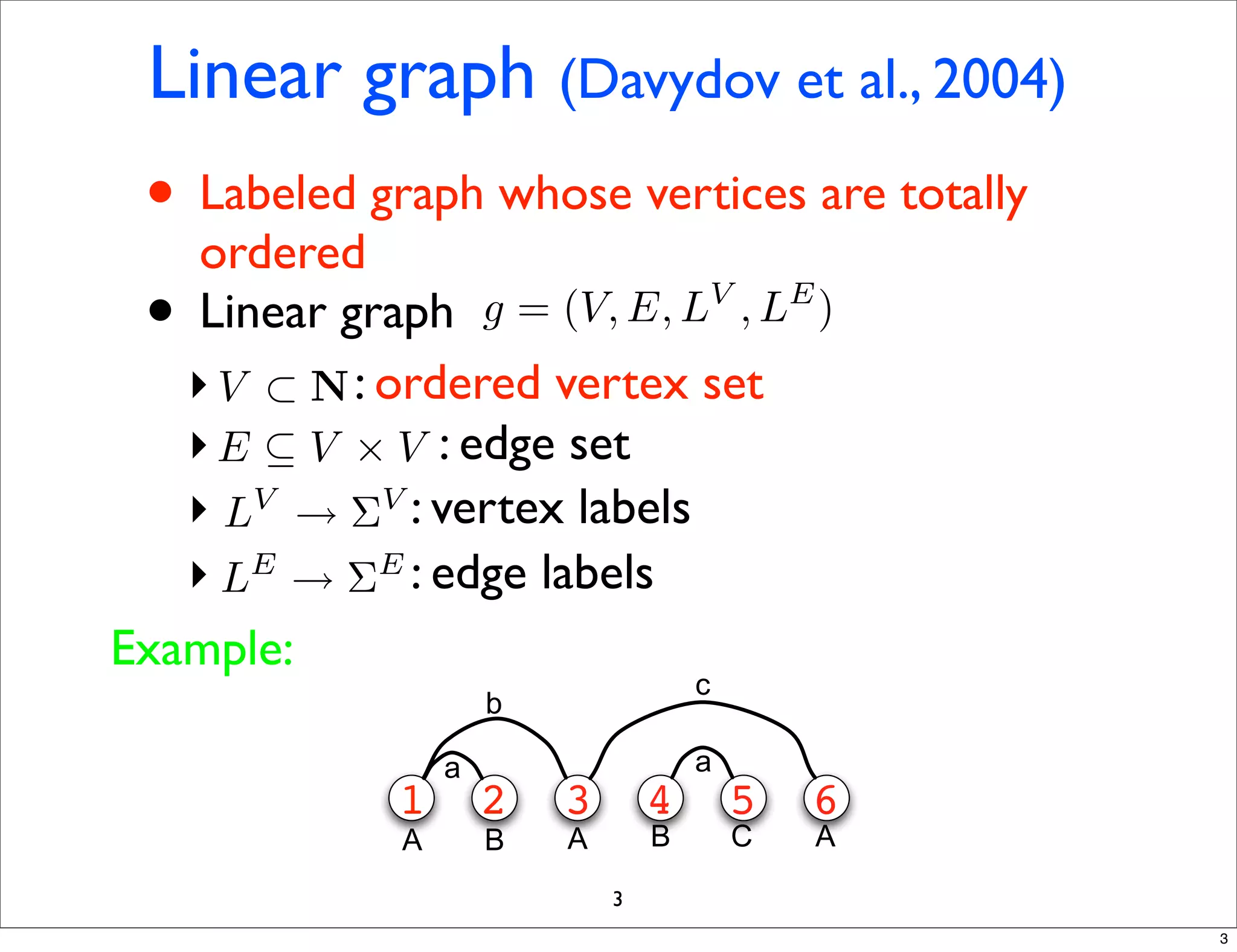
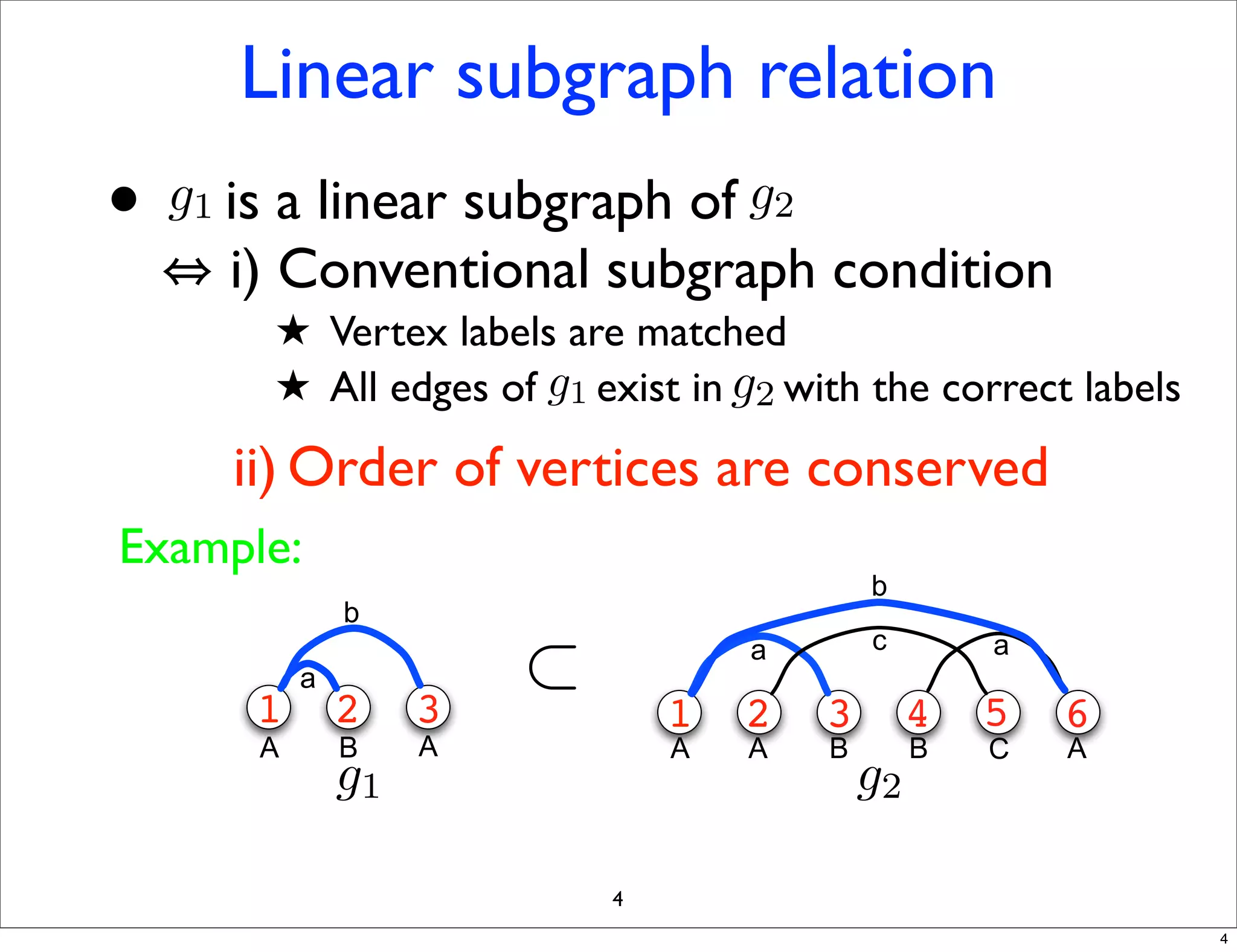
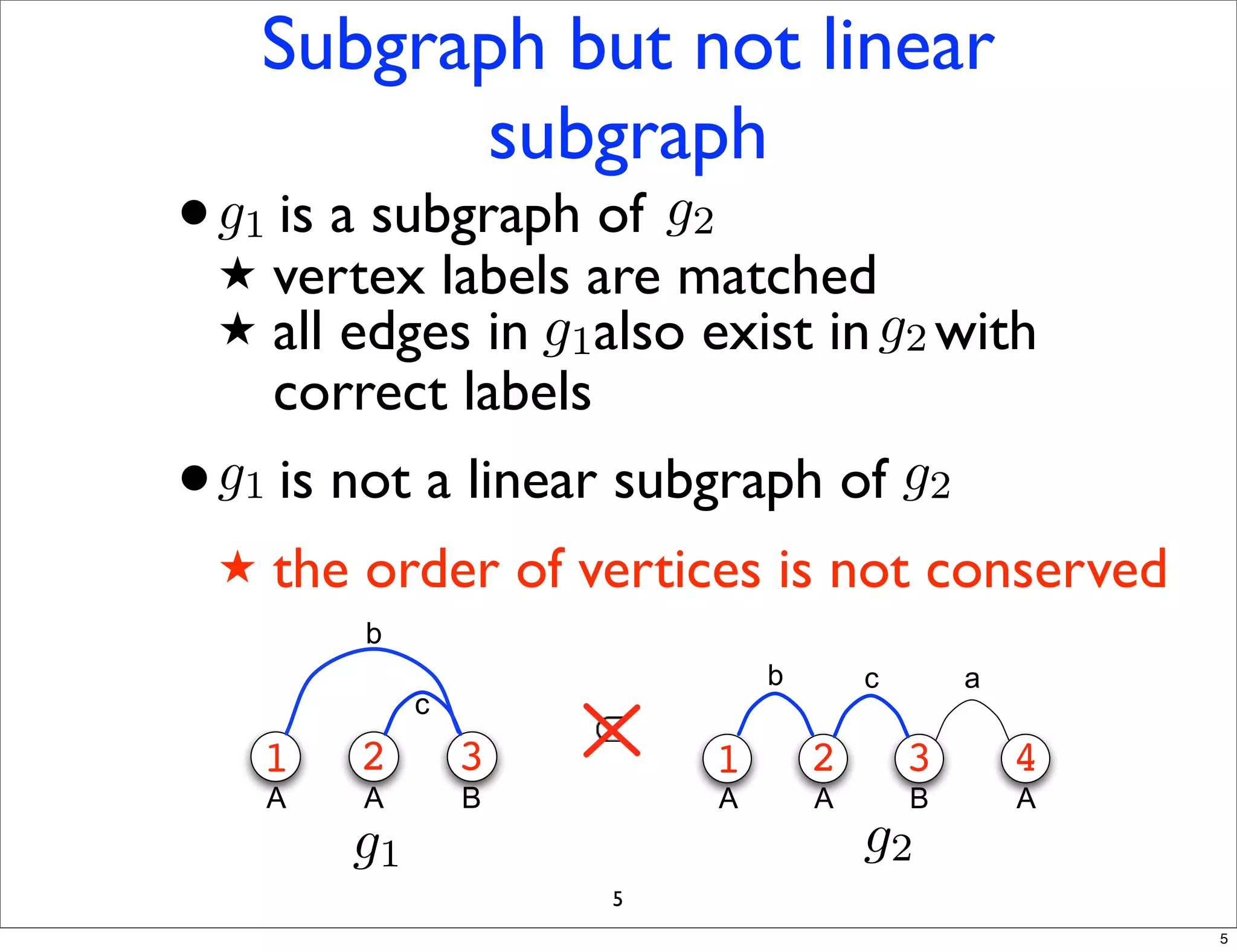
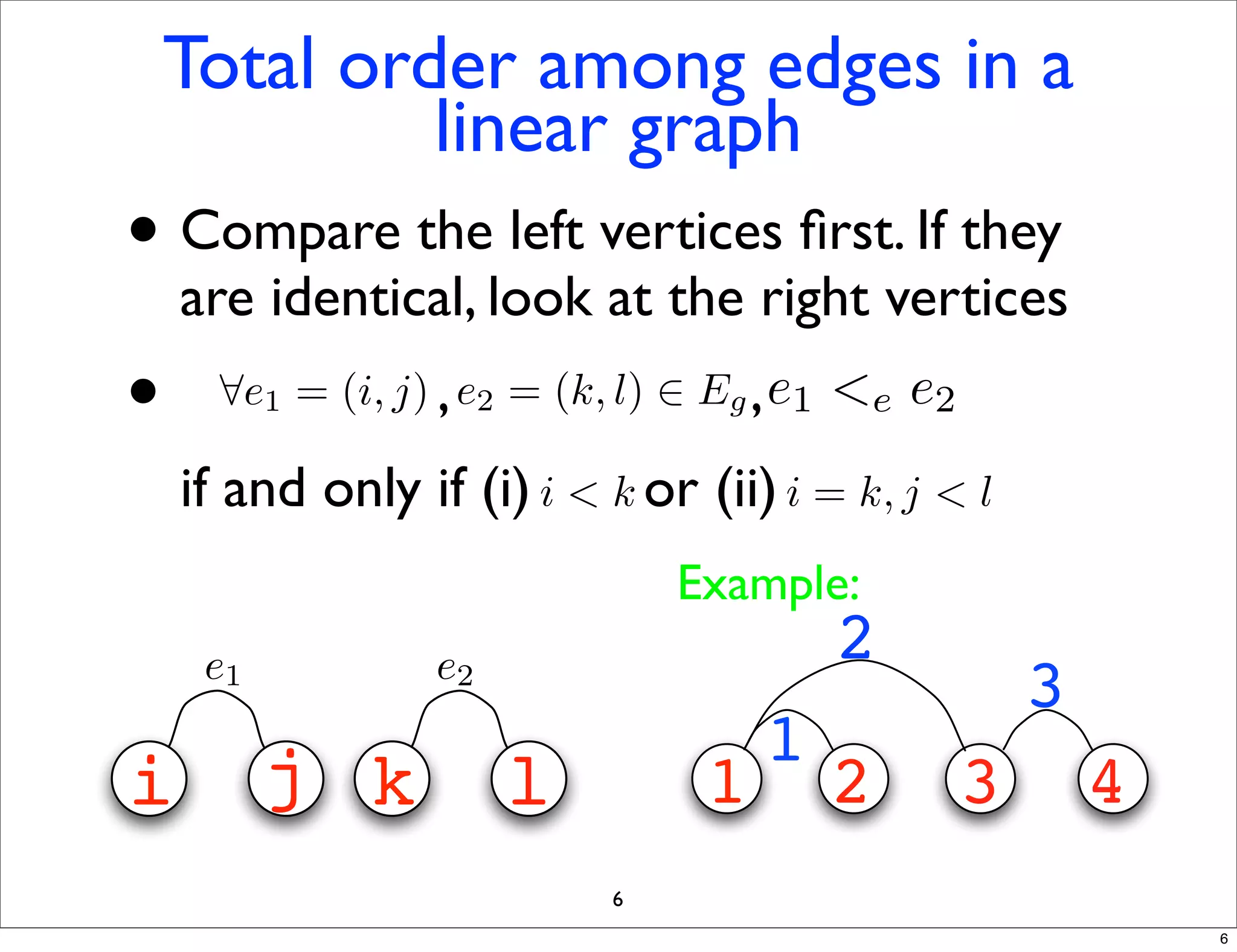
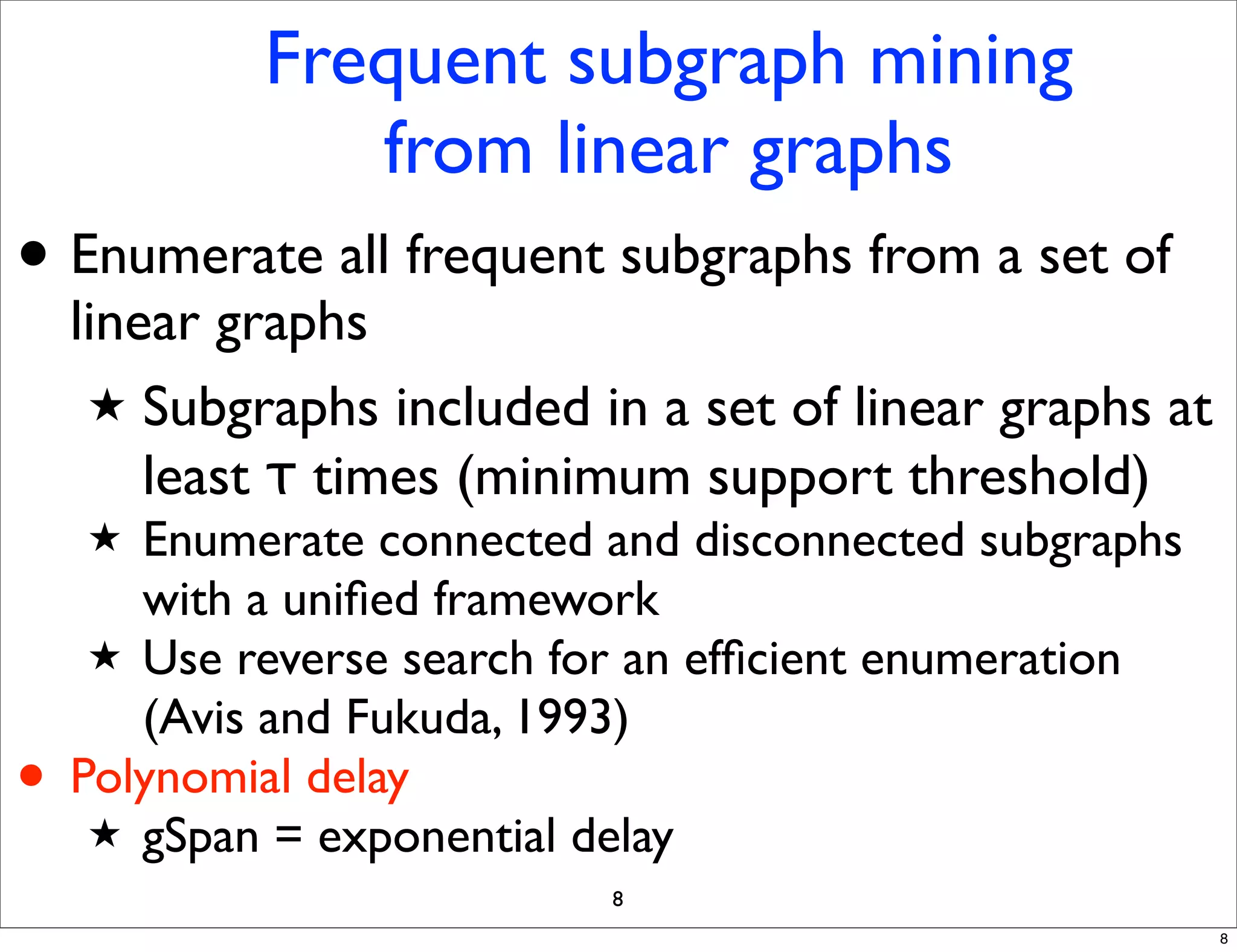
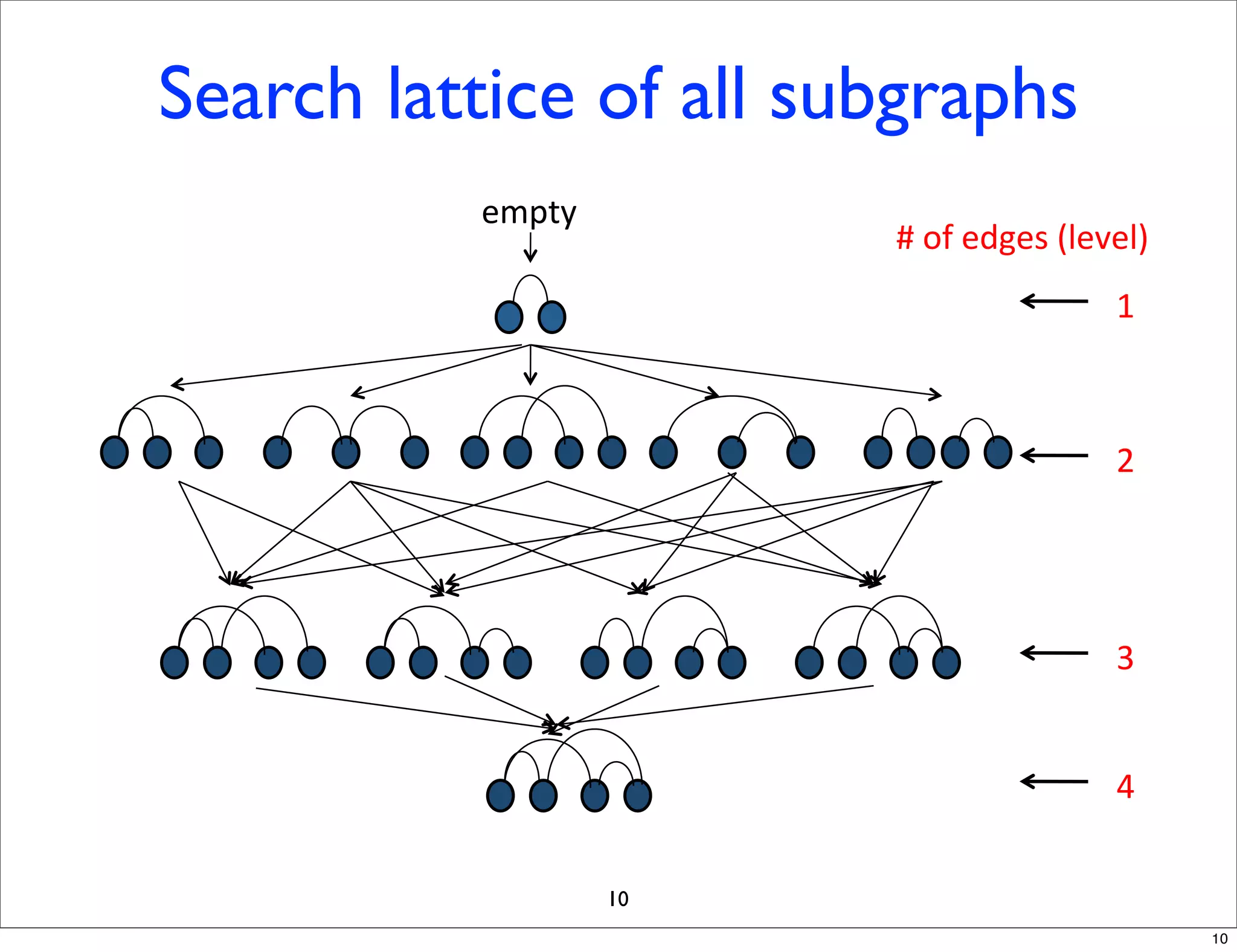
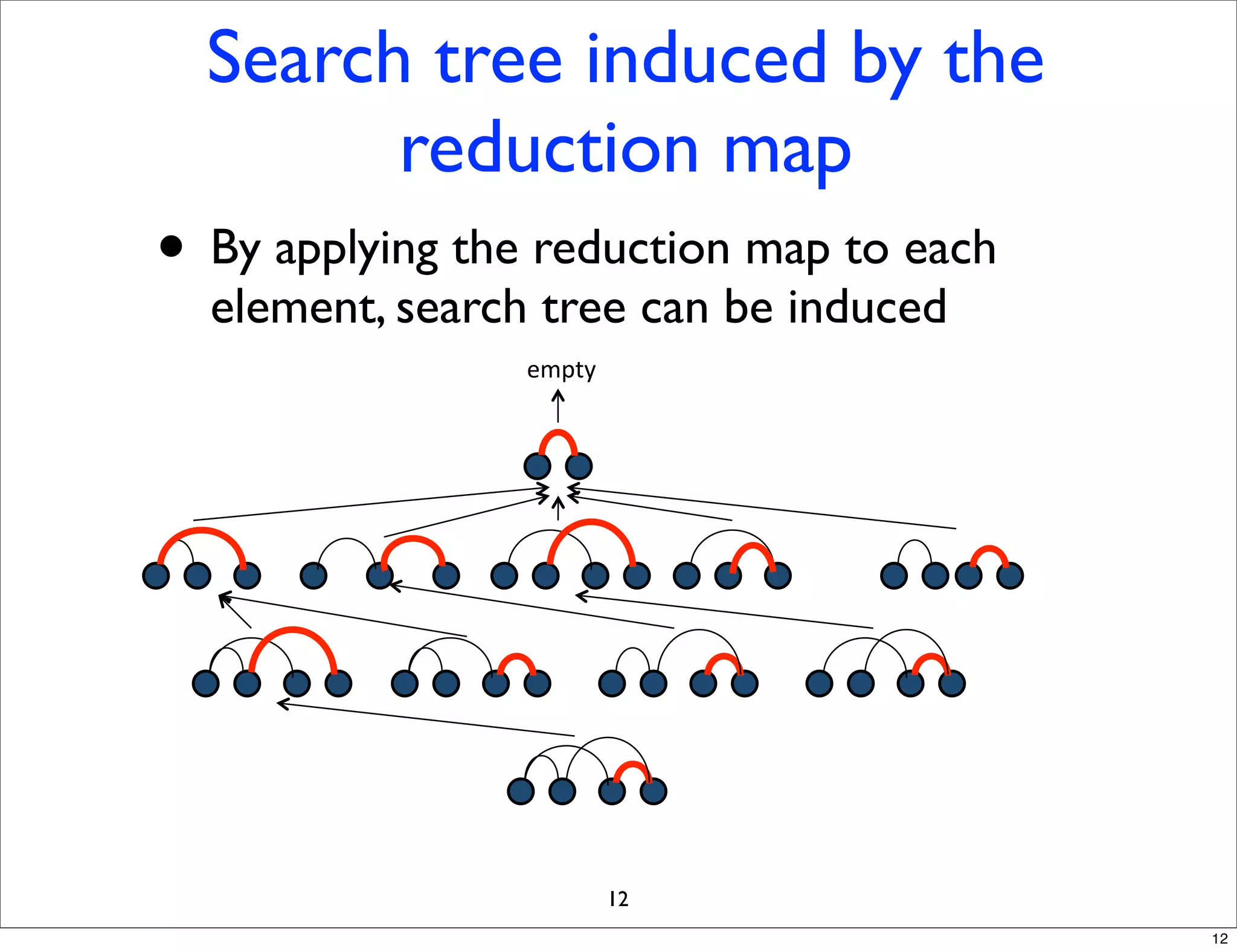
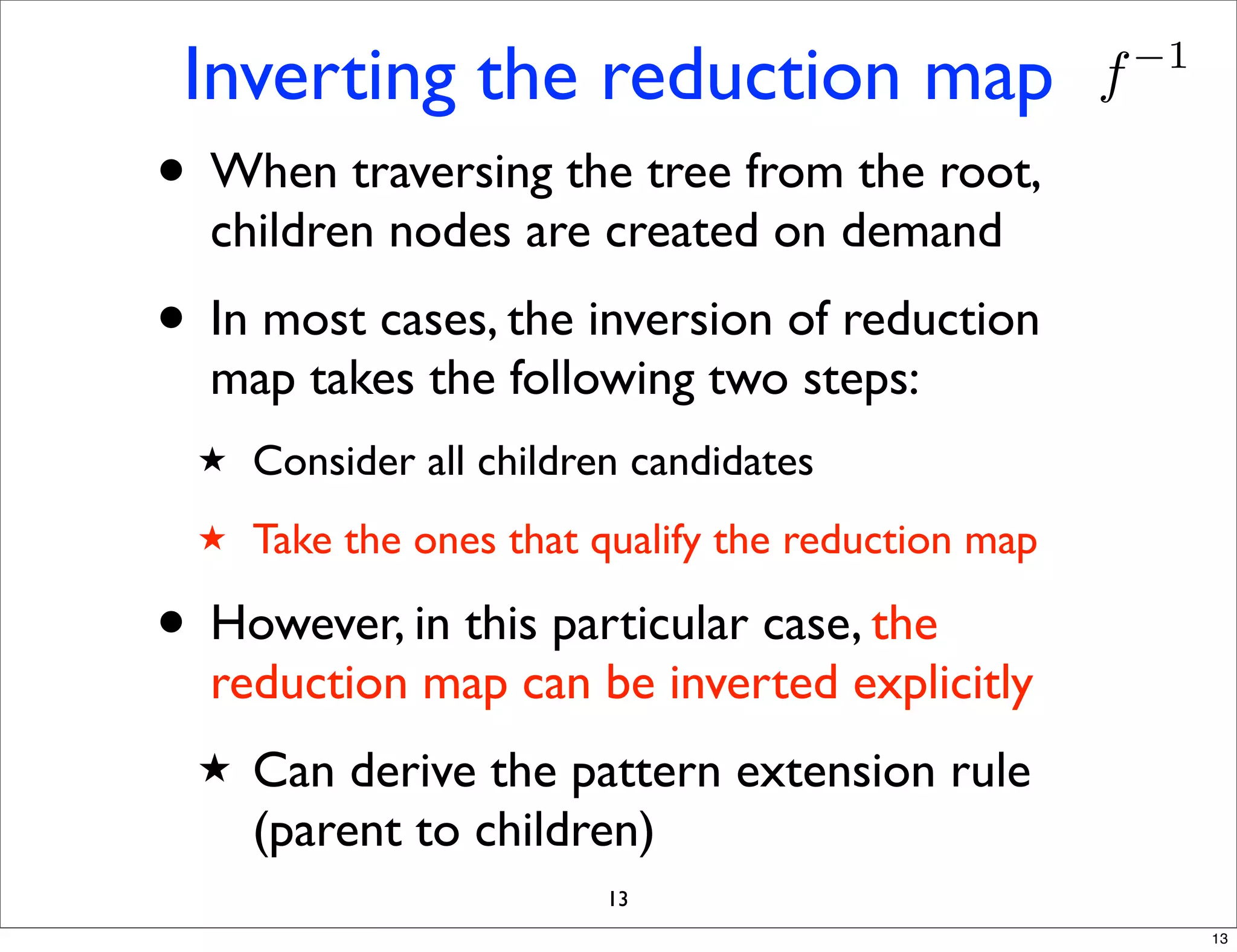
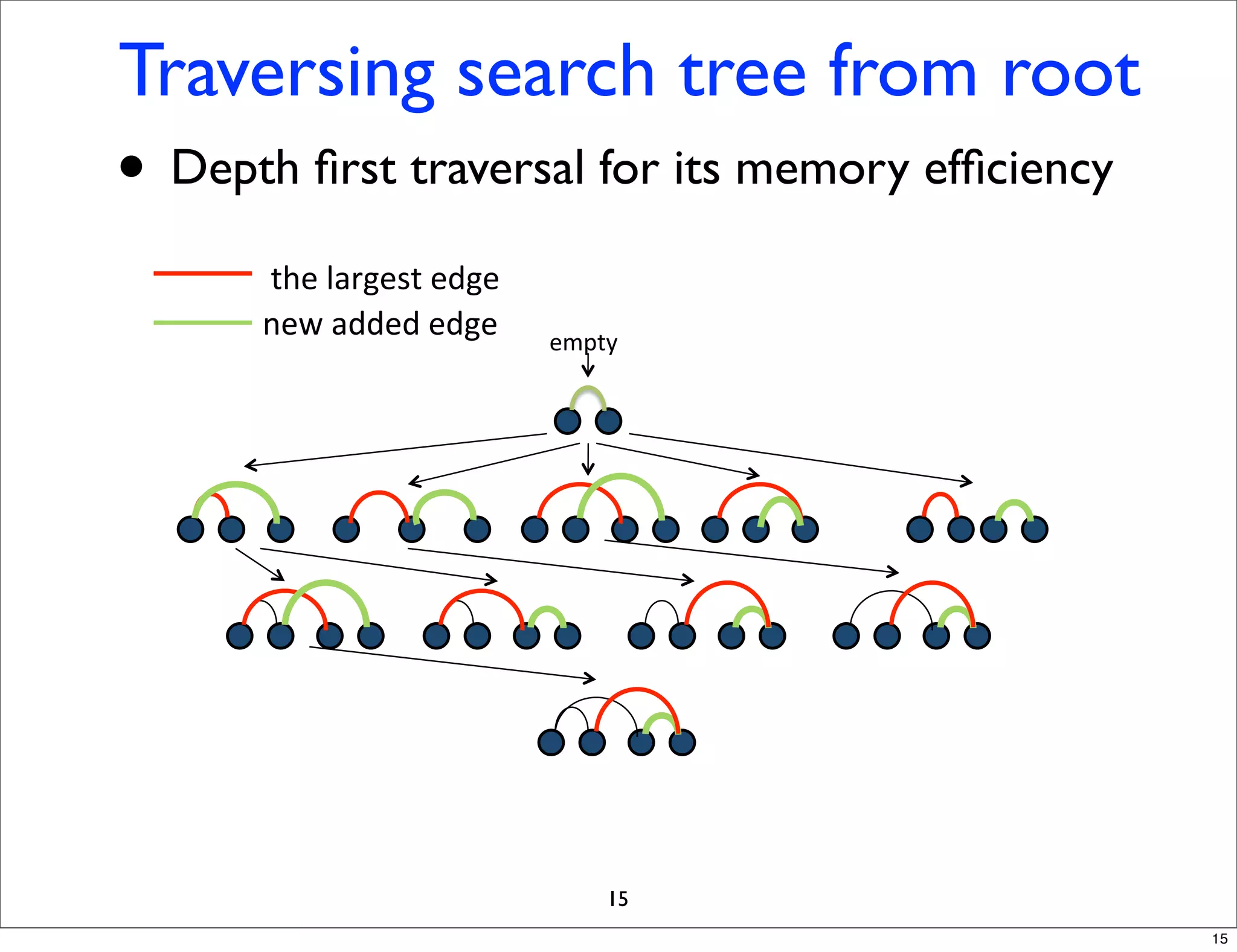
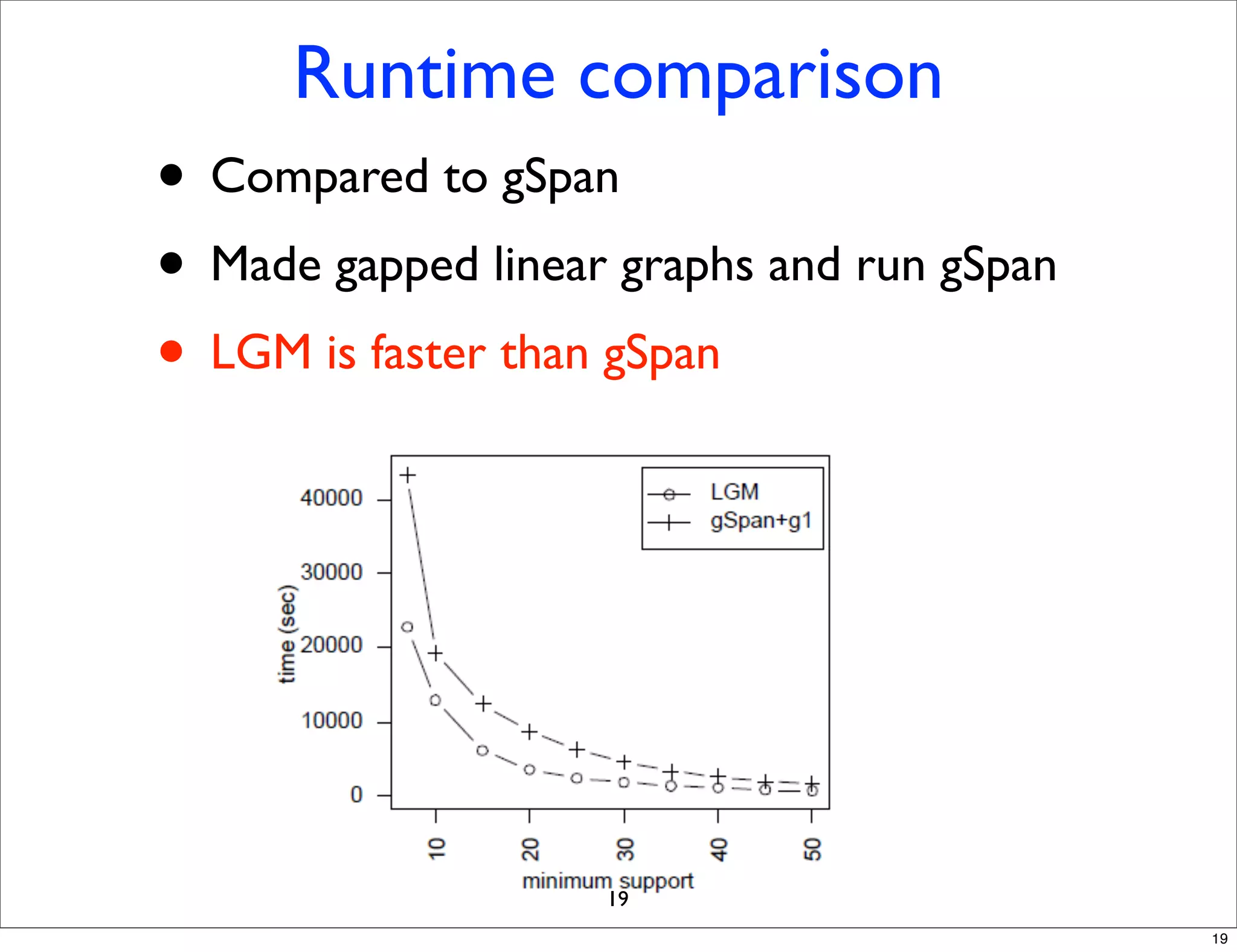
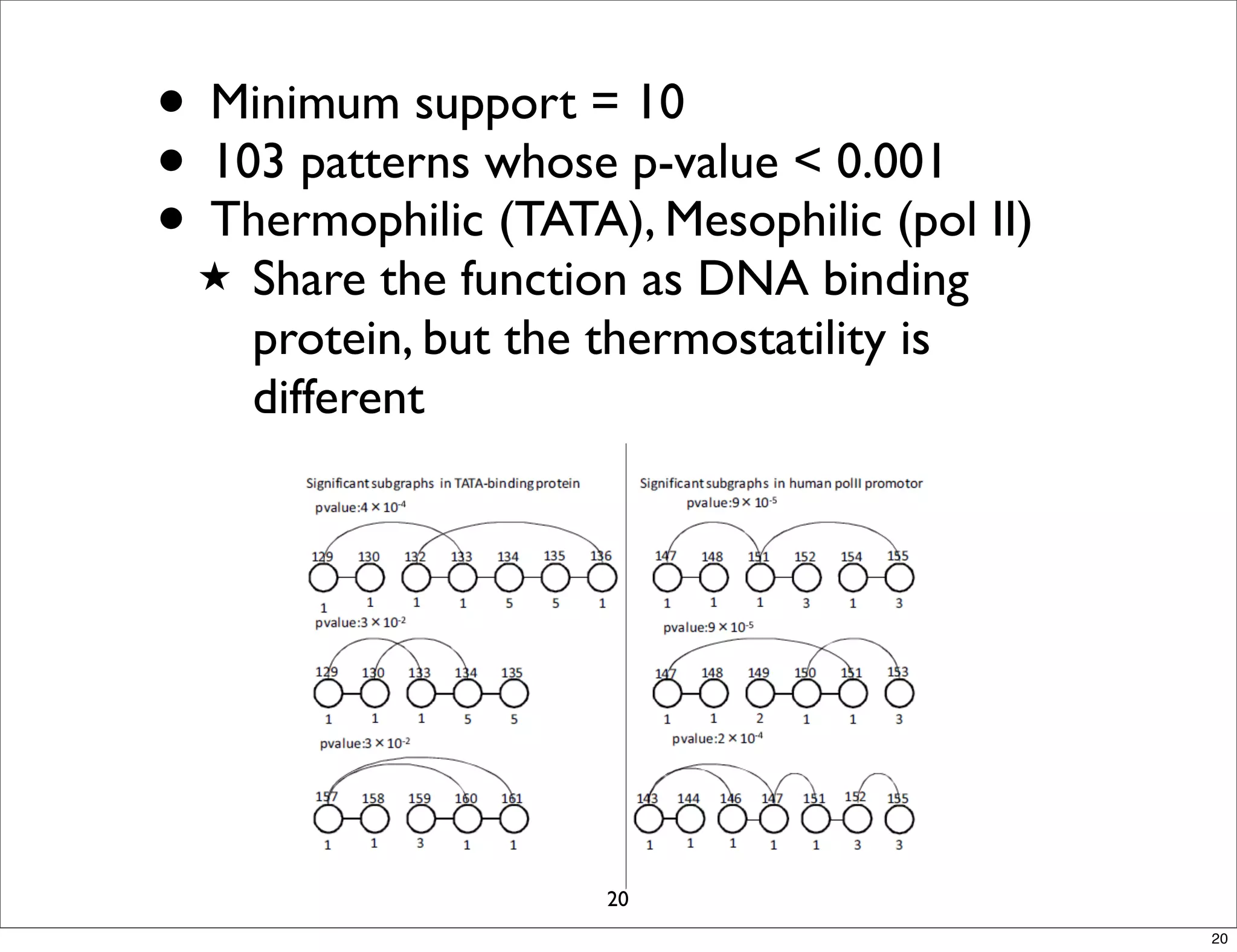
LGM is an algorithm that efficiently mines frequent subgraphs from a set of linear graphs. It uses a reverse search approach to enumerate all subgraphs without duplication, defining a search tree with a reduction map. By inverting the reduction map, it can extend patterns from parent to children nodes. Experiments apply LGM to mine motifs from protein structures, finding statistically significant patterns associated with thermophilic or mesophilic functions.