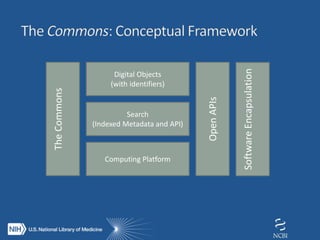
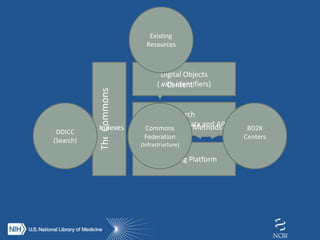
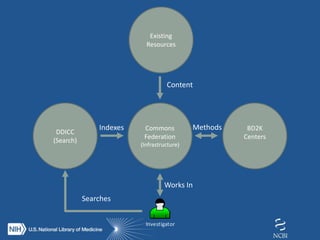
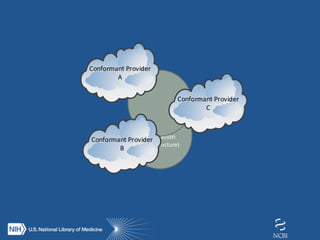
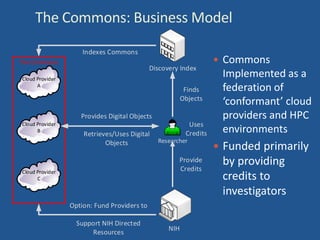
The document proposes the creation of a federated cloud computing platform called "The Commons" to support biomedical data sharing and analysis across multiple cloud providers. Key points:
- The Commons would index metadata and digital objects across conformant public and private cloud providers.
- It would be funded by providing credits to investigators for storage and computing, creating competition among providers to offer better services at lower costs.
- A phased implementation is outlined to initially involve experienced users and later expand to all NIH grantees.