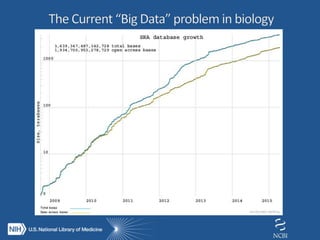
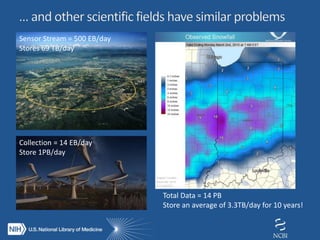
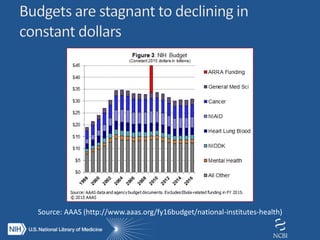
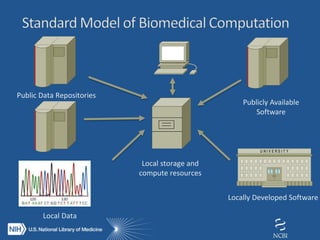
The document outlines the challenges and solutions related to managing vast amounts of biomedical data, emphasizing the need for improved infrastructure and cost-effective access for researchers. It discusses the NIH's initiative to democratize access to data and computational tools through a cloud model that supports data sharing and lowers redundancy. A pilot program will be launched to test this model, which aims to enhance data sharing, reduce costs, and improve resource management in the scientific community.




















![The modern five-line staff was first
adopted in France and became almost
universal by the 16th century
(although the use of staffs with other
numbers of lines was still widespread
well into the 17th century).
-Wikipedia
Guido [d’Arezzo 992-1050 CE] used
the first syllable of each line, Ut, Re,
Mi, Fa, Sol and La, to read notated
music in terms of hexachords; they
were not note names, and each could,
depending on context, be applied to
any note…
Much early music has been “lost” because there was no
standard representation, and the metadata for the non-
standard representations were lost (or never recorded).](https://image.slidesharecdn.com/thefutureofthecommons-151119015334-lva1-app6891/85/The-future-of-the-commons-25-320.jpg)




