JBEI Research Highlights - April 2017
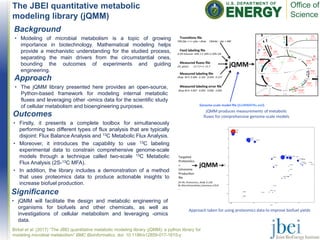
- 1. The JBEI quantitative metabolic modeling library (jQMM) Birkel et al. (2017) “The JBEI quantitative metabolic modeling library (jQMM): a python library for modeling microbial metabolism” BMC Bioinformatics, doi: 10.1186/s12859-017-1615-y Background • Modeling of microbial metabolism is a topic of growing importance in biotechnology. Mathematical modeling helps provide a mechanistic understanding for the studied process, separating the main drivers from the circumstantial ones, bounding the outcomes of experiments and guiding engineering. jQMM produces measurements of metabolic fluxes for comprehensive genome-scale models Approach taken for using proteomics data to improve biofuel yields Approach • The jQMM library presented here provides an open-source, Python-based framework for modeling internal metabolic fluxes and leveraging other -omics data for the scientific study of cellular metabolism and bioengineering purposes. Outcomes • Firstly, it presents a complete toolbox for simultaneously performing two different types of flux analysis that are typically disjoint: Flux Balance Analysis and 13C Metabolic Flux Analysis. • Moreover, it introduces the capability to use 13C labeling experimental data to constrain comprehensive genome-scale models through a technique called two-scale 13C Metabolic Flux Analysis (2S-13C MFA). • In addition, the library includes a demonstration of a method that uses proteomics data to produce actionable insights to increase biofuel production. Significance • jQMM will facilitate the design and metabolic engineering of organisms for biofuels and other chemicals, as well as investigations of cellular metabolism and leveraging -omics data.
- 2. Lipid engineering reveals regulatory roles for membrane fluidity in yeast flocculation and oxygen-limited growth Degreif et al. (2017) “Lipid engineering reveals regulatory roles for membrane fluidity in yeast flocculation and oxygen-limited growth” Metab Eng, 41, 46-56. doi: 10.1016/j.ymben.2017.03.002 Background • Yeast synthesize unsaturated lipids using the lipid desaturase OLE1 in order to maintain membrane fluidity • OLE1 expression is regulated by membrane ordering sensors in the ER, Spt23p and Mga2p; Ole1p activity is oxygen dependent • Flocculation is the clumping of yeast cells together due to cell wall interactions; it is an important phenotype in industrial fermentation Approach • OLE1 expression was titrated using the MET3 promoter and changed in methionine concentration, generating a strain that allows to functional characterization of lipid unsaturation in yeast • Various aspects of cell physiology (growth, fermentation), membrane properties (membrane fluidity), and gene expression (global RNA-Seq and targetd qRT-PCR) were correlated as a function of changes in lipid composition (unsaturation) Outcomes • Low lipid unsaturation triggers yeast to flocculate, which results from the membrane fluidity sensor Spt23p being a strong regulator FLO1, a mediator of yeast flocculation • RNA-Seq reveals that Spt23p and Mga2p regulate hundreds of other genes in yeast’s response to hypoxia Significance • Membrane fluidity in the ER regulates yeast flocculation during fermentation via the effects on changing oxygen concentrations on unsaturated lipid synthesis. A host of other genes involved in metabolism and physiology are regulated similarly. A B D 250 Δflo1Δflo8 PMET3 -OLE1 grown in 250 µM met Man Glc Gal Mal C18:1 shaking medium change E 250 [met] (µM) 200 150 100 50 0 - BY4742 - PMET3 -OLE1 0.0 0 20 40 60 80 100 CoA-S O CoA-S 9 10 O NADH + H+ NAD+ O2 2 H2 O OLE1 14 15 16 17 18 [met]retention time (min) C14:1 C16:1 C16:0 C18:1 C18:0 C14:0 (myristoleic acid) (myristic acid) (palmitoleic acid) (palmitic acid) (oleic acid) (stearic acid) 10 mM 320 µM 160 µM 10 µM %acylchainsunsaturated DC A B D p. 250 [met] (µM) 200 150 100 50 0 C A C 13xmyc N C High UFA Low UFA p90p90 FLO1 OLE1met ER membrane myc Spt23p Repression of the lipid desaturase OLE1 (top) using the MET3 promoter causes yeast to flocculate with increasing methionine concentrations (bottom), reflecting a decrease in unsaturated lipid content. Mechanistic model for this process involves activation of the ER membrane ordering sensor Spt23p.
- 3. Isopentenol(mg/L) 0 300 600 900 1200 Growth Rate (/h) 0.0 0.2 0.5 0.7 0.9 R² = 0.7226 475 WT 0.4 WT Figure 6 AtoB HMGS HMGR MK R74HIDI MtIPK K22M WT S208E Figure 2b R74H WT K22M S208ESlower = less Faster = more 2.4-fold Titer improvementtion activity. At such high concentrations of MVAP in vivo, the rate enhancements on kcat/ KM or kcat observed in vitro for select mutants would not necessarily be observed. Considering the importance of the native mevalonate pathways for isoprenoid metabolism, the activity of PMDsc might betightly regulated by MVAPconcentrations in vivo since a low KI would significantly decrease the rate of decarboxylation as excess MVAP accumulates. This directs MVAP flux through the phosphomevalonate kinase (PMK) enzyme to generate MVAPP, the preferred native substrate for PMDsc, rather than increasing futile decarboxylation of MVAP to IP. WT and four mutants, R74G, V230E, R74G-R147K-M212Q and R74H-R147K-M212Q, werespecifically selected for substrate inhibition study, where activity wasmeasured in wide range of MVAPconcentra- tions up to 100 mM (Fig. 7). These variants representatively spanned a wide range of isopentenol titers (8 – 1080 mg/ L), and their substrate inhibition behavior was characterized by determining each mutant's KI (Table 3). Interestingly, two higher isopentenol producing mutants (R74G and R74H-R147K-M212Q) exhibited significantly higher KI's (110 mM and 80 mM, respectively) than those with lower isopentenol producers (WT, 18 mM; V230E, 10 mM; and R74G-R147K-M212Q, 11 mM) (Fig. 7). A correlation emerged between growth rates (and/ or titers) and PMD activity when an observed turnover number was calculated at 100 mM MVAP using the analytical expression for noncompetitive substrate inhibition, kobs (Supplementary Fig. S7). This turnover number (kobs) depends upon each mutant's kcat, KM, and KI, perhapsyielding amoreaccurate assessment of theturnover conditions in vivo (Kang et al., 2016). Therelationship between growth and kobs of these mutants suggests that the screening platform directly reports on the extent of substrate inhibition in PMDsc mutants (Fig. 7 and Supplementary Fig. S7). This sensitivity of an in vivo screening platform to a mutant's KI modulation speaks to its power in analyzing the robustness of heterologously expressed mevalonate pathways with- in overexpressing bacterial strains. 3. Conclusion Evaluation of enzyme li a screening method. In this because it does not require designs with the desirabl essential for growth of the study, wedesigned agrowt decarboxylation activity to achieve thisgoal, the grow decarboxylation rate of M formation of IP is ultimate MVA pathway (Fig. 1A). C mutagenesis generated two which were tested by our growth-based screening pla that significantly increased L. Correlation of growth ra PMD mutants confirmed platform, and kinetics stud of this screening platform b to the target enzymeactivit Alteration of PMD and IPK inhibition strength of fosm with more flexibility, pot isopentenol production via 4. Materials and method 4.1. Plasmids and Strains All plasmids and str Supplementary Table S1. and PMD variants are show 4.2. Development of thePM For the screening platf HMGR, HMGS, MK, Idi a constructed by adding Idi for isopentenol production while HMGR, HMGS, and archaeal IP kinases from (MTH) and Thermoplasma a plasmids, pET15b-MTH a Poulter, 2010), using p IPKTHA-F-BglII and IPKTH coding apotential IPkinase two primersEcIPK-BglII-Fa Wild type PMDsc and PMD plasmid under control of a based plasmid under contr et al., 2011). Mutant libra Table 3 Analysis of Ki and kobs for select mutants. N.D.: not detected. kcat, sec− 1 KM, mM Ki, mM kobs, sec− 1 WT 0.15 2.3 18 0.02 (0.01) R74G 0.14 3.4 110 0.07 (0.04) V230E 0.07 0.8 10 0.006 (0.004) R74G-R147K-M212Q 0.22 0.5 11 0.022 (0.008) R74H-R147K-M212Q 0.16 0.43 80 0.07 (0.03) Fig. 7. Determination of the noncompetitive substrate inhibition constant, KI, for select PMDsc mutants: WT (circles); R74G(squares); V230E (upward triangles); R74G: R147K: M212Q (downward triangles); R74H: R147K: M212Q (diamonds). All activities reported are normalized relative to each mutant's kcat derived from fits to the noncompetitive substrate inhibition expression. Parameters were derived from three technical replicates (n = 3). A. Kang et al. 132 Higher KI High-throughput enzyme screening platform for the IPP-bypass mevalonate pathway for isopentenol production Kang et al. (2017) "High-throughput enzyme screening platform for the IPP-bypass mevalonate pathway for isopentenol production" Metab Eng. 41:125-134, doi:10.1016/j.ymben.2017.03.010 Background • Isopentenol (or isoprenol, 3-methyl-3-buten-1-ol) is a drop-in biofuel and a precursor for commodity chemicals such as isoprene. To overcome limitations of the original mevalonate pathway for isopentenol production such as high ATP demand and IPP toxicity, we developed an “IPP-bypass” isopentenol pathway using the promiscuous activity of a mevalonate diphosphate decarboxylase (PMD). However, the promiscuous PMD activity toward non-native substrate was still limiting and it is important to improve the enzyme activity to apply this new pathway for high titer, rate, yield production. Approach • We developed a high-throughput screening platform that correlated promiscuous PMD activity toward MVAP with E. coli cell growth. By inhibiting all IPP production from the endogenous MEP pathway, we made the heterologous pathway which depends on promiscuous PMD activity the only pathway to produce IPP which is required for the E. coli growth. Outcomes and Significance • Design of growth-linked screening platform to improve promiscuous activity of PMD. • Identification of PMD mutants that improve the conversion of MVAP to IP. • 2.4-fold improvement of isoprenol titer via the IPP-bypass pathway with PMD mutants. • Kinetic study of PMD mutants to elucidate modes of action for the titer improvement.
- 4. Impact of lignin polymer backbone esters on ionic liquid pretreatment of poplar Kim et al. (2017) “Impact of lignin polymer backbone esters on ionic liquid pretreatment of poplar” Biotechnol Biofuels, 10(1), 101. doi, 10.1186/s13068-017-0784-2 Background • Biomass pretreatment remains an essential step in lignocellulosic biofuel production, largely to facilitate the efficient removal of lignin and increase enzyme accessibility to the polysaccharides • There have been significant efforts in planta to reduce lignin content or modify its composition to overcome the inherent recalcitrance Approach • In this work, transgenic poplar lines in which monolignol ferulate conjugates were synthesized during cell wall development to introduce, during lignification, readily cleavable ester linkages into the lignin polymer backbone (i.e., “Zip lignin” produced by John Ralph at GLBRC) were pretreated with different ionic liquids (ILs). Outcomes • The strategic introduction of ester bonds into the lignin backbone resulted in increased pretreatment efficiency and released more carbohydrates with lower energy input. • After pretreatment with any of three different ILs, the transgenic poplars, especially those with relatively higher amounts of incorporated monolignol ferulate conjugates, yielding up to 23% higher sugar levels compared to wild-type plants. Significance • This work clearly demonstrate that the introduction of ester linkages into the lignin polymer backbone decreases biomass recalcitrance in poplar has the potential to reduce the energy and/or amount of IL required for effective pretreatment. • This result could enable the development of an economically viable and sustainable biorefinery process
- 5. Structure of aryl O-demethylase offers insights into its tyrosine-dependent catalytic mechanism Outcomes • Solved the X-ray crystal structure of LigM at 1.81-Å resolution (Fig 1) and showed it has a novel protein fold. • Characterize aryl O-demethylation reaction as having an ordered sequential mechanism. • Computationally determined the positions of the substrates and showed Tyr247 acts a general acid in the reaction mechanism. Kohler et al. (2017) “Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism” Proc. Natl Acad. Sci. USA, 114(16), E3205–E3214. http://doi.org/10.1073/pnas.1619263114 Background • Some strains of soil and marine bacteria have evolved intricate metabolic pathways for metabolizing environmentally-derived aromatics. • Many of these metabolic pathways go through intermediates such as vanillate. • Demethylation of vanillate is essential for downstream assimilation in the TCA cycle. • LigM from Sphingomonas paucimobilis is a novel demethylase biochemical assays. Significance • First structural and mechanistic characterization of single-domain aryl demethylase using a novel tyrosine-dependent mechanism. • Insights from this work will inform synthetic biology approaches to convert underutilized aromatics into higher value compounds. Approach • We studied the structure – function relationship of LigM using X-ray crystallography (@ALS), molecular simulations and enzymology (@JBEI). Figure 1. The 1.81 Å X-ray crystal structure of LigM reveals a novel demethylase fold and well-conserved folate-binding architecture. A) Overall structure of LigM showing both vanillate and tetrahydrofolate in the active sit where –CH3 is transferred from vanillate to tetrahydrofolate during the reaction. B) Zoomed in view highlighting the the tyrosine (Tyr247) dependent mechanism. Figure 2. LigM utilizes an ordered, sequential kinetic mechanism. The steady-state kinetics for LigM O- demethylation of vanillate were determined by varying the concentration of one substrate while the concentration of the other substrate was fixed at different concentrations.
- 6. Transport of the essential cell wall precursor UDP-arabinofuranose into the Golgi of plants Rautengarten et al. (2017) “The elaborate route for UDP-arabinose delivery into the Golgi of plants” Proc. Natl Acad. Sci. USA 114, 4261–4266, doi: 10.1073/pnas.1701894114. Background • Arabinose is a key component of plant wall polymers and glycoproteins. The majority of Ara found in glycans occurs as a furanose ring (Araf), while its precursor has a pyranose ring configuration (UDP-Arap). • The biosynthesis of UDP-Arap originates from UDP-xylose in the Golgi lumen, subsequently UDP-Arap is transported out of the lumen into the cytosol where it is converted by a mutase into UDP-Araf. a) Plants overexpressing UDP-Araf transporters showed clear developmental and growth defects, and b) The amount of Araf in total cell wall material from plants overexpressing UDP-Araf transporters demonstrated increase total arabinose content, thus indicating that the phenotype could be related to the amount of arabinose in the cell wall. In vitro assay demonstrating the transport of UDP-Araf by candidate transporters. Approach • Given the partitioned biosynthesis of UDP-Ara species, it was envisioned that a transporter capable of transporting UDP-Araf into the Golgi apparatus should exist. • Employing the state-of-the-art transporter assay developed at JBEI, we screened the entire family of Arabidopsis nucleotide sugar transporters using UDP-Araf as a substrate. Outcomes • We were able to identify a small family of four transporters with the ability to efficiently transport UDP-Araf under in vitro conditions. • By manipulating the expression levels of the transporters, we were able to demonstrate that plants with elevated expression can have increased Araf content in cell wall polymers which are accompanied by developmental defects. Significance • The identification of four UDP-Araf transporters has confirmed the circuitous route of this important cell wall substrate from the Golgi to the cytosol and back into the Golgi.
- 7. Treatment of lignite and thermal coal with low cost amino acid based ionic liquid-water mixtures To et al. (2017) "Treatment of lignite and thermal coal with low cost amino acid based ionic liquid-water mixtures" Fuel, 202, 296-306, https://doi.org/10.1016/j.fuel.2017.04.051. Background • Coal remains a significant source of energy worldwide, and efficient pre-processing of coal can significantly enhance efficiency. • The objective of this study is to explore the use of cheaper ionic liquids; made from simple acid-base reactions between two naturally occurring chemicals, choline and amino acids; for the pretreatment of lignite coal. 5x Microscopic profile of lignite and thermal coal soaked in [Ch][Lys] on a heated stage at 70OC. Darkening of the solution and swelling of the particles can be observed. Approach • Two types of coal were investigated in this study, a lignite coal with particle size 20 mesh (841 µm), and a thermal bituminous coal with particle size 75-120 µm. • Treatment of the two types of coal in the ionic liquids were carried out at a 1:5 weight/volume ratio i.e. 0.1 g of coal in 0.5 mL of ionic liquids, at 70o C with stirring for 3 hours. Outcomes • The treatment of lignite using [Ch][ARG], [Ch][GLY] and [Ch][PHE], reduces the size of the lignite particles 3-5x. • The removal of oxygenated functional groups is a desirable outcome, since oxygen reduces the efficiency of converting coal to fuels/chemicals. Significance • Treatment of coal by ionic liquids reduces the decomposition temperature of coals, the size of the coal particles, and removes oxygenated functional groups, all of which are beneficial to downstream combustion of coal.
- 8. Preliminary crystallographic analysis and in vitro characterization of the nonmutase UAM2 from Oryza sativa Outcomes • Preliminary crystallographic analysis we performed as well as analysis of the oligomeric state in crystal and solution. UAM2 crystallizes readily but forms crystals of limited quality. • We show by size exclusion chromatography that the nonmutase is monomeric in solution, and employ limited proteolysis to show DTT-enhanced proteolytic digestion, indicating the existence of intramolecular disulphide bridges that can protect the protein from proteolysis. Welner et al. (2017). "X-ray diffraction analysis and in vitro characterization of the UAM2 protein from Oryza sativa" Acta Crystallogr F Struct Biol Commun, 73(Pt 4), 241-245, doi: 10.1107/S2053230X17004587 Background • The maintenance of UDP-activated sugars in plant cells is important for the synthesis of complex carbohydrates including cellulose and hemicelluloses. • The existence of seemingly non-enzymatic proteins in complexes interconverting UDP-arabinopyranose and UDP-arabinofuranose in the plant cytosol remains an enigma. Significance • A better understanding of the underlying molecular mechanism and pathways involved in the maintenance of activated sugar pools in plant cells will help in the engineering of bioenergy feedstocks Approach • To shed light on their function, we have undertaken crystallographic and functional studies of the nonmutase UAM2 from Oryza sativa. Crystals of OsUAM2. The largest dimension is approximately 100 µm. Size exclusion chromatogram of OsUAM2 (blue), overlaid with molecular weight standards (red)
- 9. Leveraging microbial biosynthetic pathways for the generation of ‘drop-in’ biofuels Background • As the majority of petroleum consumption in the United States consists of gasoline (47%), diesel fuel and heating oil (21%), and jet fuel (8%), ‘drop- in’ biofuels that replace these petrochemical sources are particularly attractive. • With our increased understanding of biosynthetic logic of metabolic pathways, we discuss the unique advantages of fatty acid, terpene, and polyketide synthases for the production of bio-based gasoline, diesel and jet fuel. Production of biofuels in E. coli from three metabolic pathways Maximum possible theoretical yield and highest titer given for each compound based on the substrate glucose under anaerobic conditions. Approach • Diverse fuel energy needs of the global economy may be best served by diversifying the metabolic pathways we choose to develop biofuel substitutes. Outcomes • Fatty acid synthesis is currently the most mature pathway for bioproduction of straight-chain hydrocarbons of short to medium chain length that compose gasoline and diesel. • Exquisite control over product formation by polyketide synthases could generate customized products with desired combustion properties as biofuel substitutes. • Monoterpenes (C10) and sesquiterpenes (C15), produced from isoprenoid synthases, are identified as diesel and jet fuel substitutes. Significance • Fatty acid synthases currently produce the highest titers, isoprenoid synthases result have higher energy density fuels, and the customizability of polyketides could allow the production of tailored biofuel products with desired physical properties. Pathway Compounds Mass yield % (g/ghexose) Highest reported titre Fatty Acid Synthesis Alkanes (C13-C17) 30.7 – 30.8% 300 mg /L [11] Alkanes (C9-C14) 30.5 – 30.7% 580 mg /L [16] Propane 29.4 % 32 mg /L [20] Isoprene Synthesis Bisabolene 32.4 % 5.2 g / L [33] Farnesene 32.4 % 1.1 g/L [36] Pinene 32.4 % 32 mg/L [38] Limonene 32.4 % 700 mg/L [46] Polyketide Synthesis Multi-methyl-branched esters 27.2 – 27.6 % 98 mg/L [51] Pentadecane 30.8 % 140 mg/L [52] Zargar et al. (2017) “Leveraging microbial biosynthetic pathways for the generation of ‘drop- in’ biofuels” Curr Opin Biotechnol, 45, 156-163, doi: 10.1016/j.copbio.2017.03.00
- 10. NH O n OH O H2N n ORF26 n=1-3 H2N O OH ORF26 E. coli H2N O NH2 H2N O OH NH2 DavB DavA NH O H2N O OH NH2 NH O Application of an acyl-CoA ligase from Streptomyces aizunensis for lactam biosynthesis Zhang et al. (2017) “Application of an acyl-CoA ligase from Streptomyces aizunensis for lactam biosynthesis” ACS Synth Biol. doi: 10.1021/acssynbio.6b00372 Background • -caprolactam and -valerolactam are important commodity chemicals used in the manufacture of nylons, with millions of tons produced annually. • Biological production of these highly valued chemicals has been limited due to a lack of enzymes that cyclize ω-amino fatty acid precursors to corresponding lactams under ambient conditions. • In this study, we demonstrated production of these chemicals using ORF26, an acyl-CoA ligase involved in the biosynthesis of ECO- 02301 in Streptomyces aizunensis (Fig. 1) Fig 1. Lactam chemistry of ORF26 and the valerolactam biosynthesis from lysine Fig 2. Production of valerolactam and caprolactam from precursor feeding Approach • To apply ORF26 for renewable chemical production, ORF26 was overexpressed in E. coli, and both valerolactam and caprolactam were formed in vivo by feeding their respective precursors, 5- aminovaleric acid (5-AVA) and 6-aminocaproic acid (6-ACA) (Fig. 2) • To achieve renewable production of valerolactam, we introduced a two-enzyme pathway into E. coli that converts lysine to 5-AVA. Outcomes • High concentrations of lysine (e.g., 10 g/L) resulted in ~5 fold increase in valerolactam production compared to non-supplemented cultures. Significance • The enzyme ORF26 enables five-membered, six-membered and even seven-membered ring formation at mild temperatures, resulting in the production of important industrial lactams such as valerolactam and caprolactam via fermentation 0 0.5 1 1.5 2 2.5 pET28-Empty pET28-ORF26 pET28-CaiC Caprolactam(mg/L) 24h 48h 72h 0 50 100 150 200 250 pET28-Empty pET28-ORF26 pET28-CaiC Valerolactam(mg/L) 24h 48h 72h H2N O OH H2N O OH ORF26 or CaiC E. coli NH O NH O O OH H2N NH O NH O O OH H2N E. coli A. B. 152.7 ± 5.4 48.6 ± 1.2 2.02 ± 0.08 ORF26 or CaiC 188.8 ± 7.5 N.D. N.D.