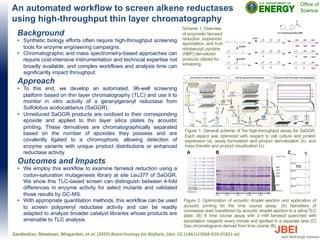
An automated workflow to screen alkene reductases using high-throughput thin layer chromatography
1) Researchers developed an automated 96-well screening platform using thin layer chromatography (TLC) to monitor the in vitro activity of an enzyme called geranylgeranyl reductase from Sulfolobus acidocaldarius. 2) The platform uses TLC to separate enzyme variants with unique product distributions or enhanced reductase activity. 3) Testing this workflow on a library of enzyme mutants, researchers could distinguish 4-fold differences in enzyme activity for some mutants and validated results with another method.