A study characterized the subcellular localizations and substrate specificities of the seven membrane-bound apyrase enzymes in Arabidopsis. It found that all seven localized internally and preferentially hydrolyzed NDPs rather than NTPs. This suggests their primary role is to efficiently convert UDP and GDP produced by glycosyltransferase reactions into UMP and GMP to regulate endomembrane nucleotide homeostasis. The study provides the first comprehensive analysis of the functions of this entire family of enzymes in a plant species.



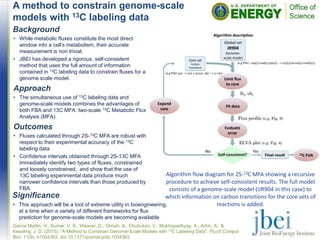
![Calorimetric evaluation indicates that
lignin conversion to advanced biofuels is
vital to improving energy yields
Background
• Biomass conversion studies have typically focused on
mass yields rather than calculating energy yields
• Typically do not capture the overall efficiencies and areas
for improving biomass conversion.
Approach
• Energy density measurements using bomb calorimetry
were applied along with mass yields to calculate energy
yields from combinations of individual processes and
lignocellulosic feedstocks using ionic liquid pretreatment.
Outcomes
• Mass yield from switchgrass (68.0% theoretical) after IL
pretreatment was lower than energy yield (61.6%
theoretical).
• In contrast, energy yield (68.0% theoretical) after IL
pretreatment of eucalyptus, was lower than MY (74.3%
theoretical)
• Due to differences in initial lignin present in the feedstock.
Gardner, J. L., He, W., Li, C. L., Wong, J., Sale, K. L., Simmons, B. A., Singh, S., & Tanjore, D.
(2015). "Calorimetric evaluation indicates that lignin conversion to advanced biofuels is vital to
improving energy yields". RSC Advances, 5(63), 51092-51101, doi:10.1039/c5ra01503k
Significance
• Strong correlation between lignin concentrations in pretreated
solids and energy densities.
• New analytical method to establish energy yield as a function of
mass yield of fermentable sugars from biomass conversion
Mass balance and energy yields from mixed feedstocks
after [C2C1Im][OAc] pretreatment and subsequent
enzymatic hydrolysis; *calculated values. Note: all energy
values are reported after adjusting energy density for ash
and moisture content in the sample.](https://image.slidesharecdn.com/jbeihighlightsseptember2015-161017213646/85/JBEI-Highlights-September-2015-5-320.jpg)
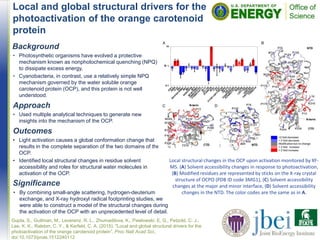


![Theoretical insights into the role of water
in the dissolution of cellulose using
IL/water mixed solvent systems
Background
• Ionic liquid (IL) pretreatment is promising, but there
remain unanswered questions into the exact
mechanisms between molecules that give rise to desired
performance during pretreatment.
Approach
• Use computational modeling (all atom MD) to
understand the fundamental interactions between water-
IL-cellulose to determine dominant forces that govern
cellulose dissolution. The IL studied is 1-ethyl-3-
methylimidazolium acetate ([C2C1Im][OAc]).
Outcomes
• Interaction of the IL [C2C1Im][OAc] and water at certain
concentrations plays as physicochemical driving forces
assisting the cellulose dissolution process.
• Preferred solvation of [OAc]- energetically favors their
accumulation around the cellulose hydroxyl groups of
the glucan chains and disrupts inter-chain H-bonds
• Increases in water concentration leads to spontaneous
solvation of both [C2C1Im][OAc] and cellulose, which in
turn disrupts the overall pretreatment process efficacy.
Parthasarathi, R., Balamurugan, K., Shi, J., Subramanian, V., Simmons, B. A., & Singh, S.
(2015). "Theoretical Insights into the Role of Water in the Dissolution of Cellulose using
IL/Water Mixed Solvent Systems". J Phys Chem B., doi:10.1021/acs.jpcb.5b02680.
Significance
• Knowledge gained from this study provides a better
understanding of the dual role played by the water (as a co-
solvent/antisolvent) in dissolving cellulose.
The final structural snapshots of the simulated system for 100 ns at 433K.](https://image.slidesharecdn.com/jbeihighlightsseptember2015-161017213646/85/JBEI-Highlights-September-2015-9-320.jpg)
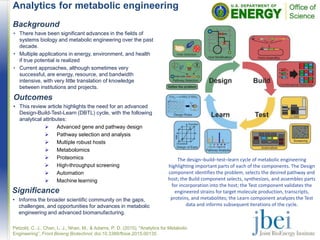
![Structural features affecting the
enzymatic digestibility of pine wood
pretreated with ionic liquids
Background
• Pinus radiata (pine) is a promising feedstock worldwode
for the production of advanced biofuels and renewable
chemicals, but is notoriously difficult to process due to
terpene content and lignin.
• Certain ionic liquids (ILs) are promising but relatively
unproven for processing pine, and the important factors
in determining sugar yields are not well known.
Approach
• Evaluate two different ionic liquids over a range of
temperatures to determine impact on structure,
chemistry, and enzyme accessibility.
Outcomes
• Detailed analysis of the substrates revealed that the
most important change brought about by the IL
pretreatments was an increase in accessible surface
area.
• Loss of cellulose crystallinity only occurred for the more
severe [C2C1Im][OAc] pretreatments.
• Maximum glucose yields of 84% were obtained.
Torr, K. M., Love, K. T., Simmons, B. A., & Hill, S. J. (2015). "Structural features affecting the enzymatic
digestibility of pine wood pretreated with ionic liquids”, Biotechnol Bioeng., doi:10.1002/bit.25831
Significance
• Provides new insights into the mechanism of IL
pretreatment and the most significant factors that
generate high yields of fermentable sugars.
Field emission scanning electron microscopy images of Pinus
radiata wood untreated (A), pretreated with [C2C1Im]Cl at 80
oC (B) and 120 oC (C), and pretreated with [C2C1Im][OAc] at
80 oC (D), 100 oC (E), and 120 oC (F). Scale bar =10 um.](https://image.slidesharecdn.com/jbeihighlightsseptember2015-161017213646/85/JBEI-Highlights-September-2015-11-320.jpg)