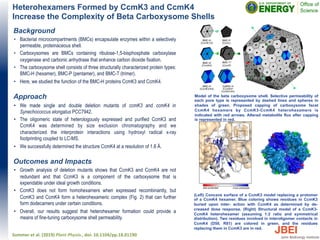
- Yeast strains from the Pfaff collection at UC-Davis were tested for their ability to produce ethanol from switchgrass hydrolysate pretreated with ionic liquids. The highest ethanol yield of 70% theoretical was achieved by Wickerhamomyces anomalus.
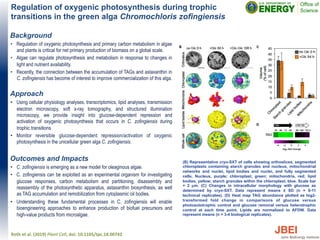
- Adding 200 mM NaCl to growth media increased E. coli growth and isoprenol production 1.5-13 fold in the presence of imidazolium-based ionic liquids. No comparable increase was observed for E. coli MG1655.
- A rice mutant (gs9-1) with shorter grain length but unchanged width was identified. Whole genome sequencing identified the gene as a new allele of gs9 involved in cellulose microf
![Ethanol production in switchgrass hydrolysate by ionic
liquid-tolerant yeasts
Background
• Ionic liquids (ILs) are a promising pretreatment for the conversion
of plant biomass to biofuels and biochemicals.
• Previous work had identified IL-tolerant yeasts in the Pfaff
collection at UC-Davis.
Approach
• IL-tolerant yeasts previously identified in the Pfaff collection were
tested for the ability to produce ethanol from switchgrass.
hydrolysate obtained by pretreatment with ILs and enzymatic
hydrolysis.
Outcomes and Impacts
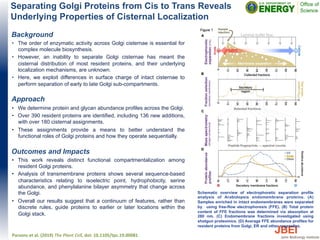
• A total of 25 yeast strains including four commercial
ethanologenic Saccharomyces cerevisiae strains were tested to
grow and ferment sugars in stepwise screening tests.
• Four yeast strains produced >10 g/L ethanol in laboratory media
containing [C2C1Im][OAc].
• The highest ethanol yield of 70% of theoretical yield was
achieved by Wickerhamomyces anomalus UCDFST 72-248
when grown in hydrolysate with 3.2% residual IL.
• This study demonstrates the value of microbial collections such
as the Pfaff collection for biotechnology application.
Sitepu
et
al.
(2019
Bioresource
Technology
Reports,
doi.org/10.1016/j.biteb.2019.100275](https://image.slidesharecdn.com/jbeihighlightsjuly2019-190821174432/85/JBEI-highlights-july-2019-1-320.jpg)
![NaCl enhances Escherichia coli growth and isoprenol
production in the presence of imidazolium-based ionic
liquids
Background
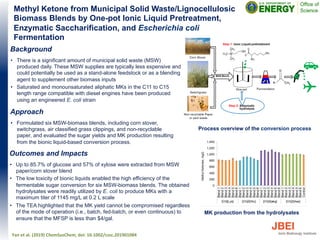
• Sustainable production of fuels and value-added chemicals from
lignocellulosic biomass in an integrated biorefinery is extremely
important for both energy security and sustainability. One of the
major steps in the conversion of biomass to fuels is pretreatment,
through which plant polysaccharides are made accessible to the
action of hydrolytic enzymes. The use of certain ionic liquids (ILs)
to process biomass is an emerging pretreatment method.
Approach
• Bacteria are intolerant to high concentrations of imidazolium-based
ILs. The effect of NaCl on IL tolerance was tested to see if
chaotropic effects might improve bacterial tolerance.
Outcomes and Impacts
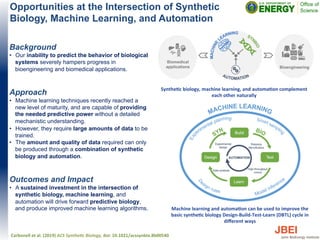
• Addition of 200 mM NaCl to growth media with 50 mM [C4C1Im]Cl
and 50 mM [C4C1Im][OAc] increased growth of for E. coli DH1 and
MG1655 2-6 fold.
• Addition of 200 mM NaCl increased isoprenol production by E. coli
DH1 1.5-13 fold in the presence of imidazolium Ils.
• No comparable increase was observed under the same conditions
for MG1655.
• The inclusion of NaCl enhances the IL tolerance of E. coli strains,
providing a complement to genetic approaches.
Wang
et
al.
(2019)
Bioresource
Technology
Reports,
doi:
10.1016/j.biteb.2019.01.021](https://image.slidesharecdn.com/jbeihighlightsjuly2019-190821174432/85/JBEI-highlights-july-2019-2-320.jpg)