This document discusses the Baltimore classification of viruses. It begins by explaining the classification was developed by David Baltimore and groups viruses based on their nucleic acid composition and replication strategy. It then provides details on each of the 7 virus groups:
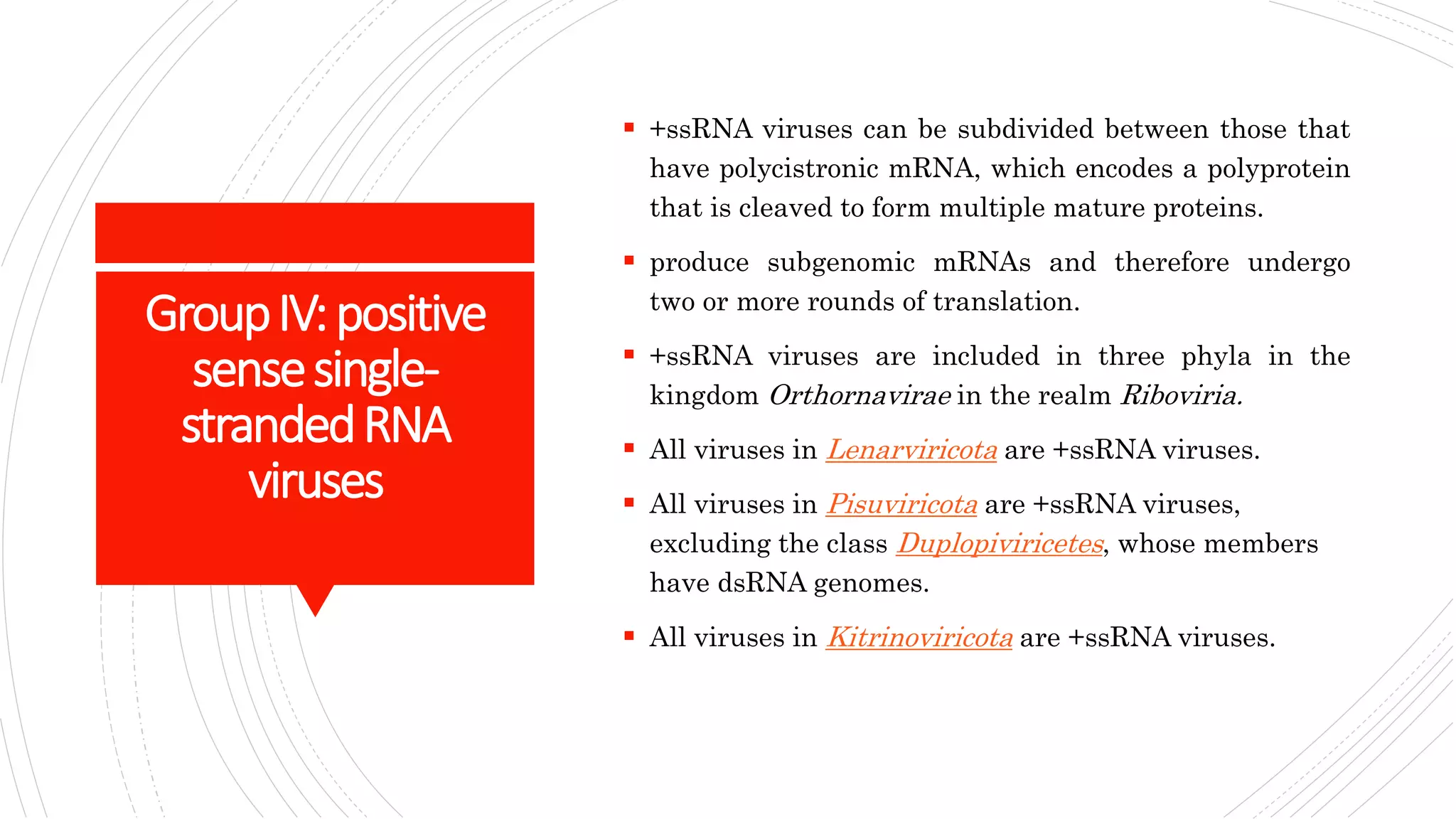
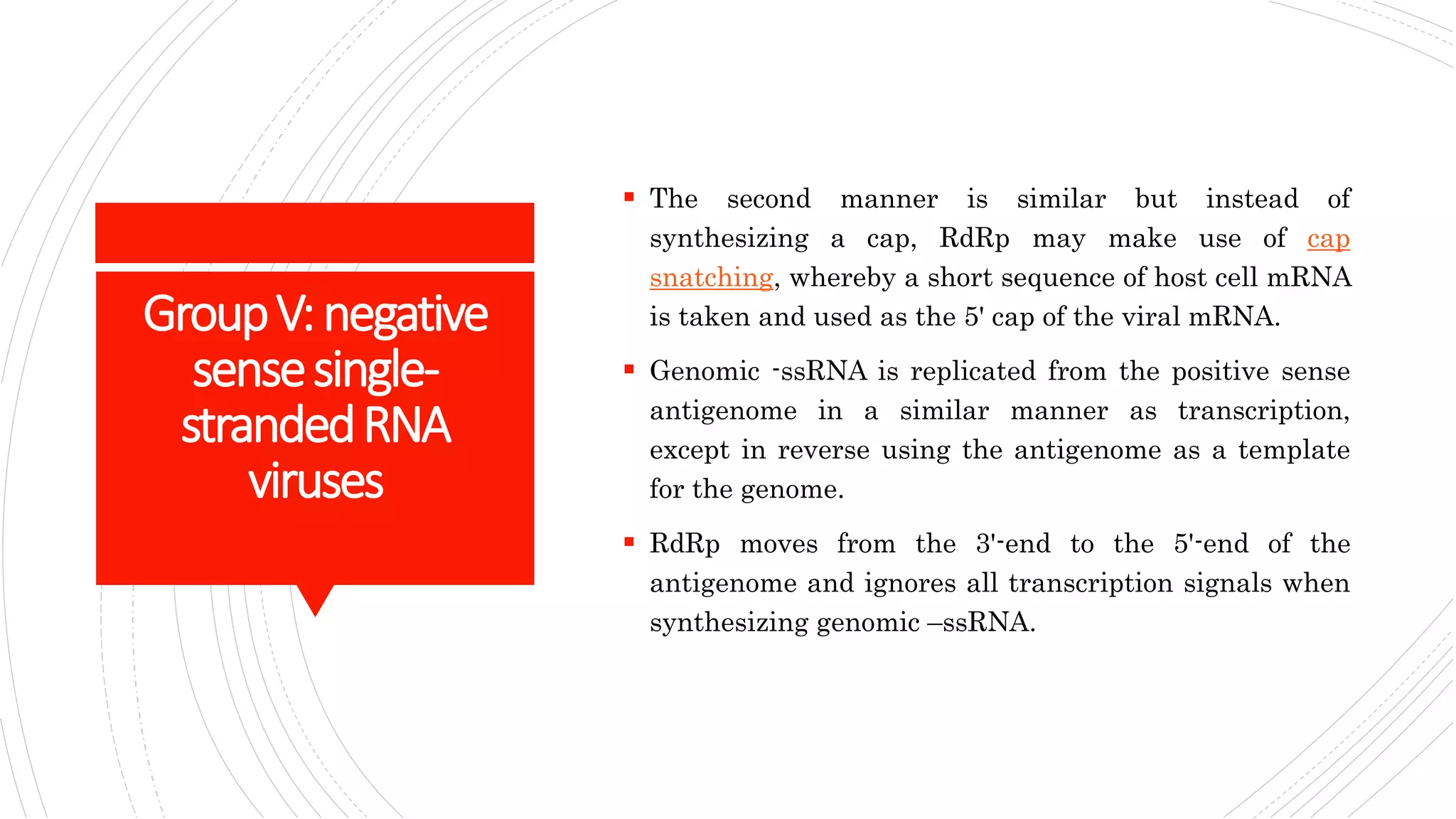
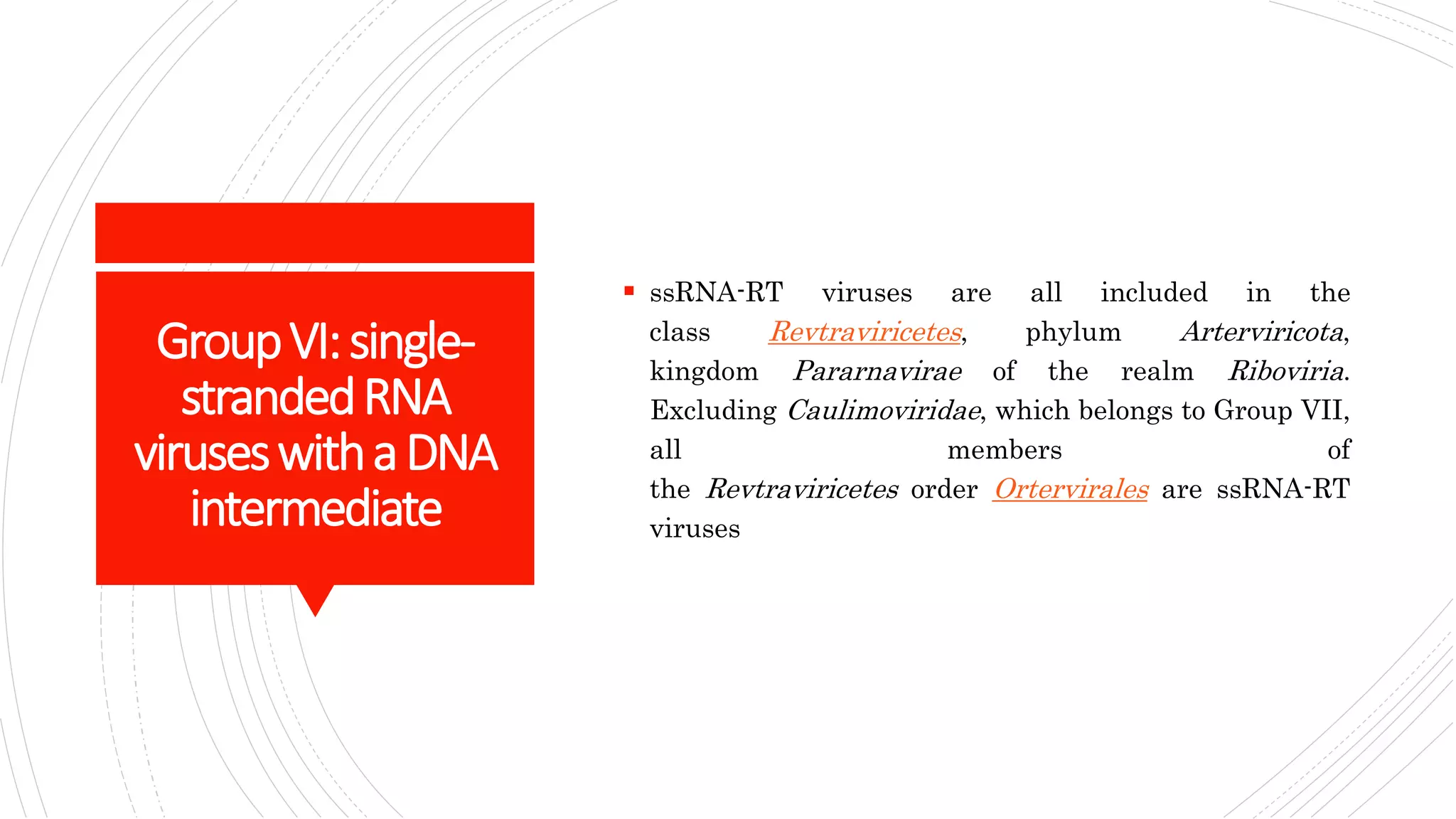
Group I contains double-stranded DNA viruses like adenoviruses and herpesviruses. Group II contains single-stranded DNA viruses like parvoviruses. Group III contains double-stranded RNA viruses like reoviruses. Group IV contains positive-sense single-stranded RNA viruses like picornaviruses. Group V contains negative-sense single-stranded RNA viruses like orthomyxoviruses. Group VI contains positive-sense single-str











![Examples
All viruses in Duplodnaviria are dsDNA viruses.
Viruses in this realm belong to two groups: tailed
bacteriophages in Caudovirales and herpesviruses
in Herpesvirales.[12]
In Monodnaviria, members of the
class Papovaviricetes are dsDNA viruses.
Viruses in Papovaviricetes constitute two
groups: papillomaviruses and polyomaviruses.
All viruses in Varidnaviria are dsDNA viruses.
Viruses in this realm include adenoviruses, giant
viruses, and poxviruses.[13]](https://image.slidesharecdn.com/introbaltimoreclassification-200827155728/75/Intro-baltimore-classification-14-2048.jpg)