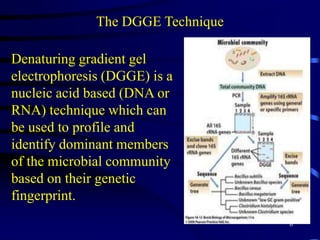
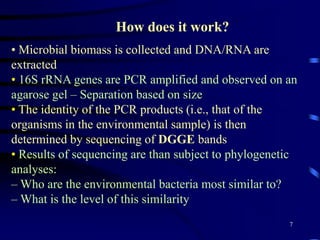
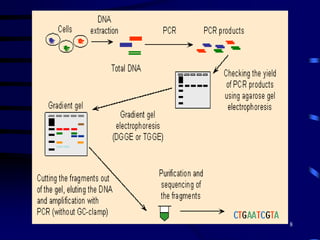
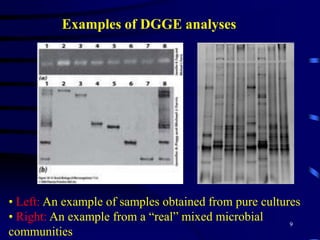
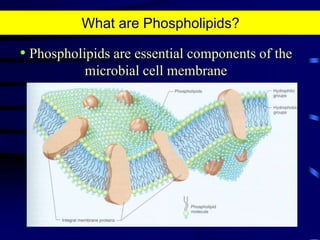
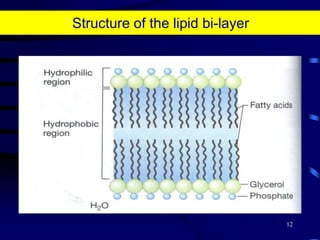
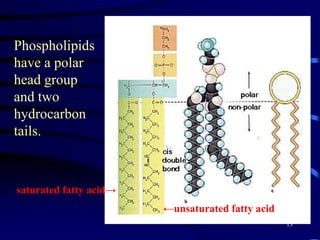
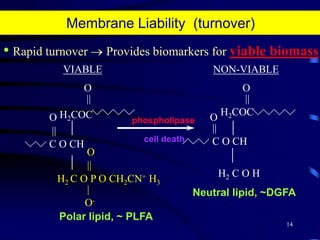
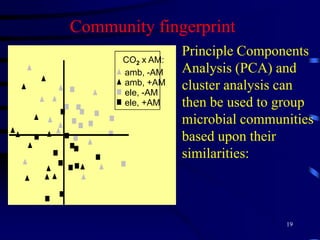
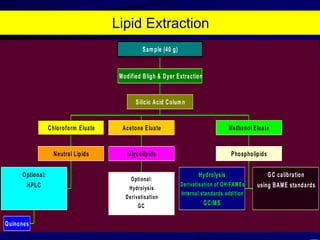
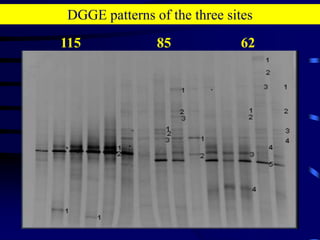
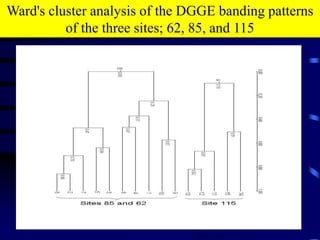
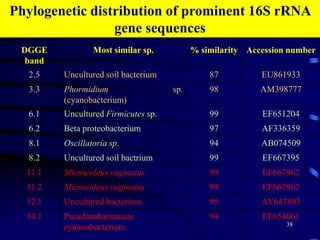
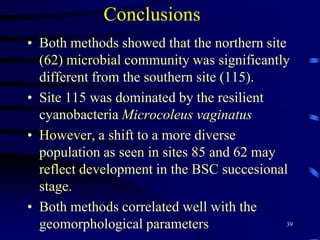
1. The document discusses two complimentary methods, DNA-based DGGE and lipid biomarker analysis, for assessing soil microbial communities.
2. It provides examples of how these methods have been used to study microbial community structure and diversity across different soil environments and conditions.
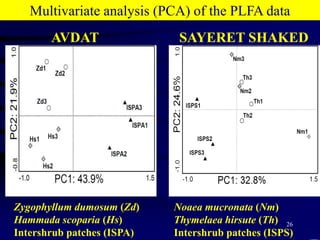
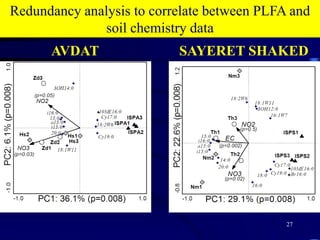
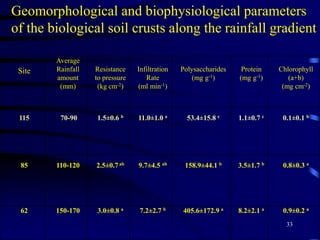
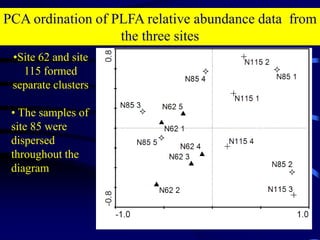
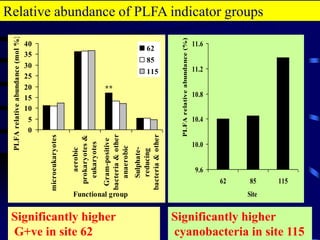
3. Specifically, it summarizes two studies that used these methods: one found evidence of "microbial diversity patchiness" correlated with vegetation in Negev desert soils, while the other found bacterial community succession along a desert rainfall gradient.