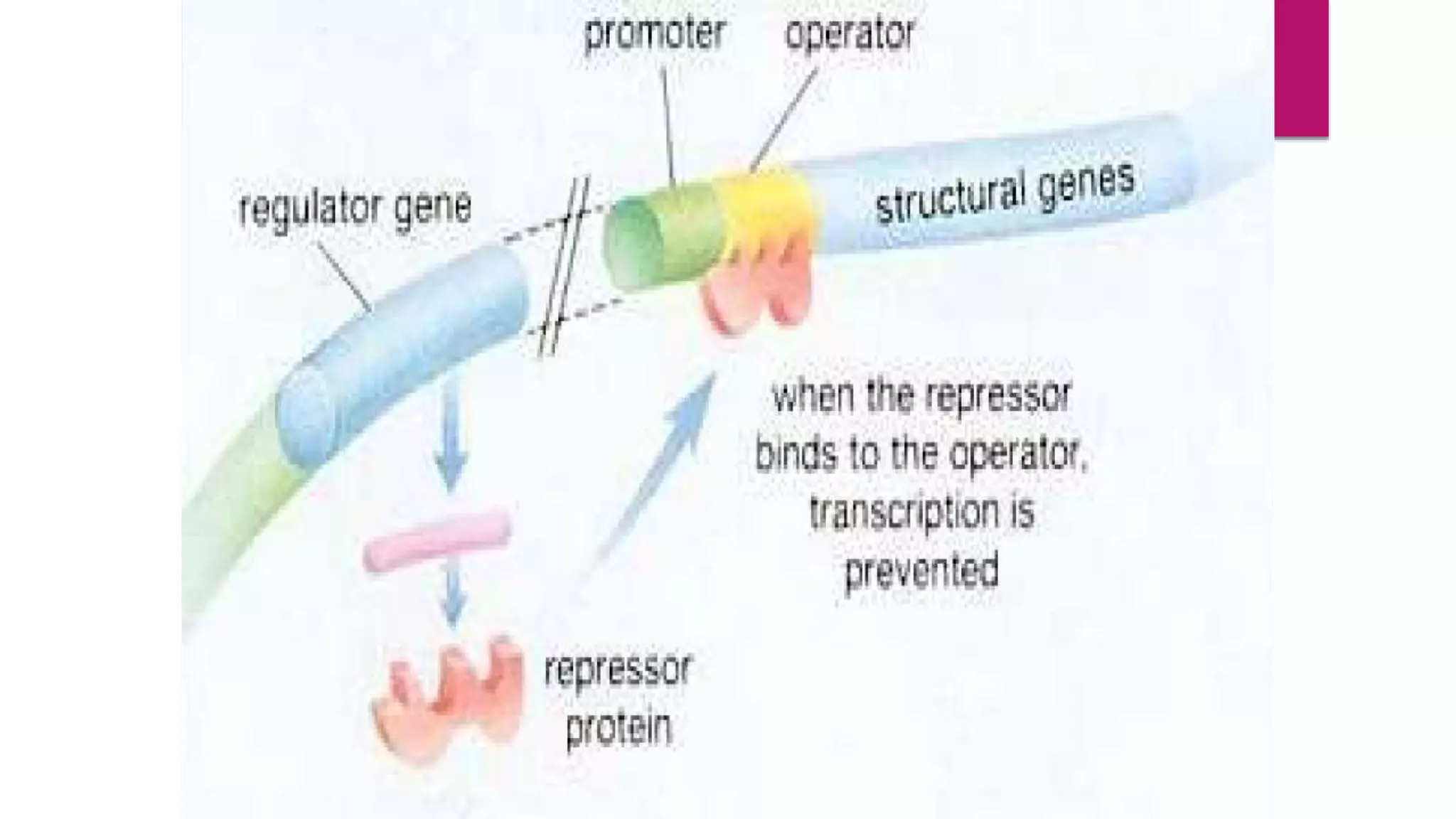
In prokaryotes, genes are regulated at the transcriptional level to control protein synthesis. There are two main types of gene regulation: negative and positive. Negative regulation occurs via a repressor protein that binds to an operator region to block transcription. Positive regulation happens when an inducer molecule activates the promoter, promoting transcription. The lac operon is an example of this, containing genes for lactose metabolism that are induced by the presence of lactose via the lac repressor protein. The operon model of gene regulation explained by Jacob and Monod helped describe this process.