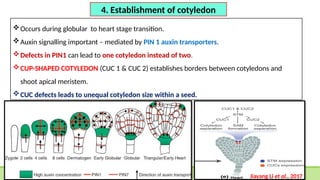
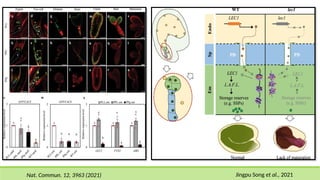
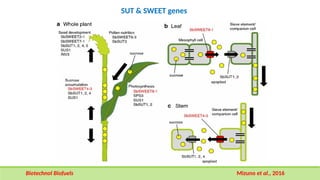
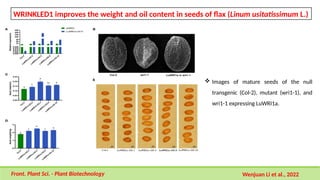
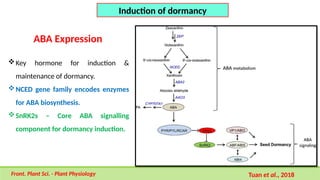
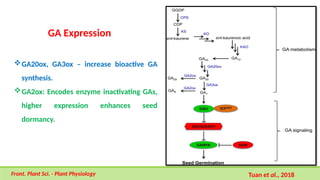
This comprehensive presentation explores the intricate genetic mechanisms controlling seed development and maturation. The content covers key stages from embryogenesis through maturation, detailing crucial molecular processes and regulatory networks. Special focus is given to important genes and transcription factors like WOX, LEC, and ABI families, along with hormone signaling pathways involving ABA and GA. The presentation explains critical processes including zygote polarization, tissue differentiation, meristem specification, and the establishment of dormancy. Supported by recent research findings and illustrations, this resource provides valuable insights into the molecular basis of seed development. Perfect for graduate students studying about seed development and maturation.