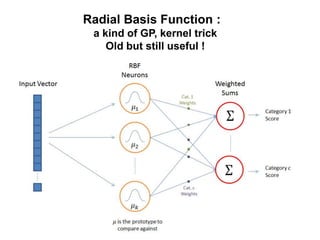
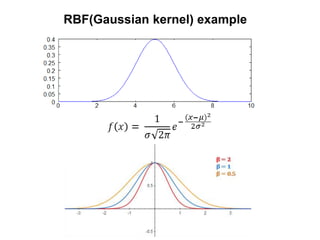
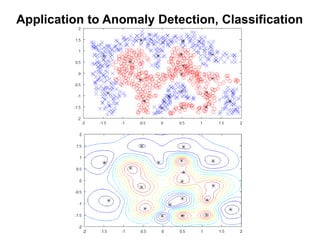
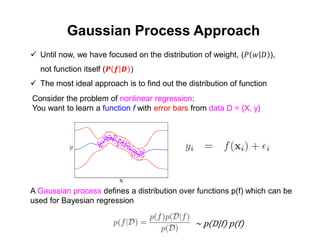
The document discusses Gaussian Processes (GPs) for regression, classification, and optimization, emphasizing their ability to provide closed-form predictions, handle small data, and remain explainable. It details how GPs place a prior over functions rather than model parameters, offering a Bayesian approach to learning functions with uncertainty. Key points include the importance of the covariance function in GP, the sampling procedure, and the implications for predictive modeling.