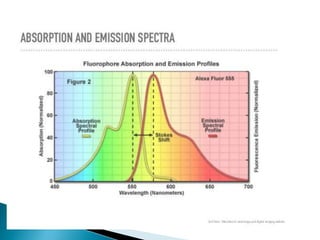
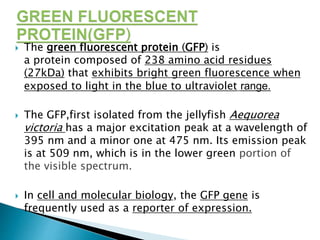
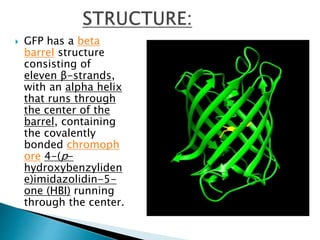
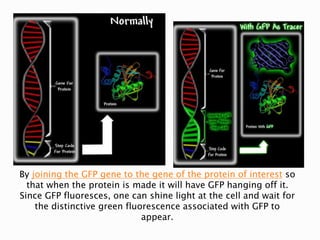
The document discusses fluorescence microscopy and fluorescent proteins. It describes how fluorescence microscopy works using light sources like mercury lamps and filters to excite and detect fluorescence. Common fluorescent proteins discussed include GFP, DsRed, and their variants. GFP derives its fluorescence from internal cyclization reactions forming a chromophore, and it and its variants are widely used as biological markers and reporters of gene expression. DsRed fluorescence comes from similar reactions and it and its variants emit light across the visible spectrum, enabling multi-color labeling experiments.