Embed presentation
Download to read offline



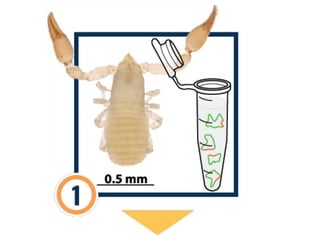
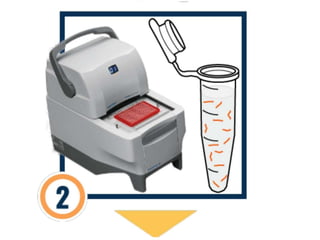
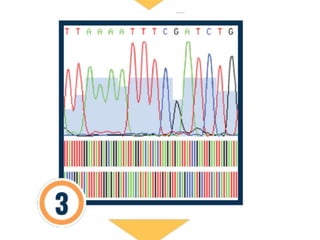



DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise is that a DNA sequence can be used to uniquely identify an organism by comparing it to a reference library of DNA sequences. For animals, the cytochrome c oxidase 1 gene is commonly used, while plants may use the rbcL, matK, or trnH-psbA genes. Fungi typically use the internal transcribed spacer region. The process involves isolating DNA from a sample, amplifying the target region via PCR, sequencing the results, and comparing the sequence to reference databases to identify the matching species. Applications include taxonomy, pest/disease control, food safety, conservation,












