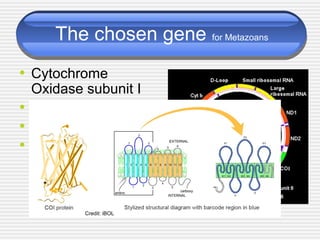
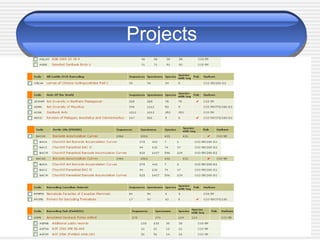
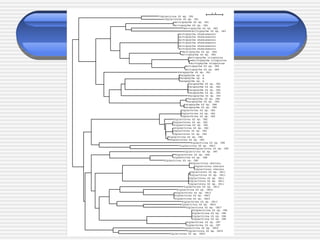
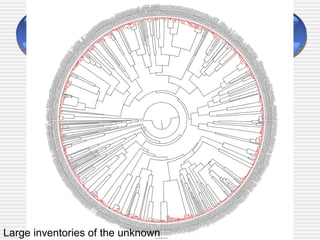
The document discusses DNA barcoding, a method for species identification based on a standardized gene region, emphasizing its importance in addressing the biodiversity crisis and aiding the discovery of new species. It outlines the applications of DNA barcoding in various fields such as food control, customs, and conservation, as well as the standards and governance necessary for effective implementation. The document highlights the necessity for taxonomic advancement through efficient identification processes and the challenges faced by traditional taxonomy.