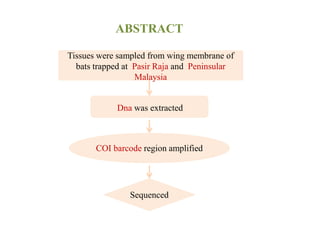
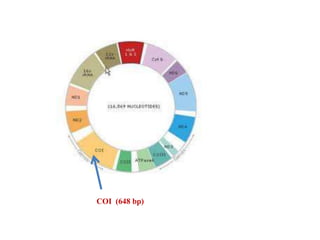
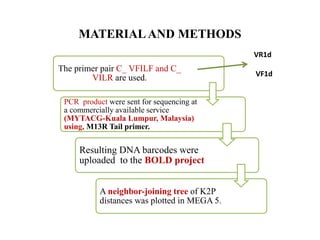
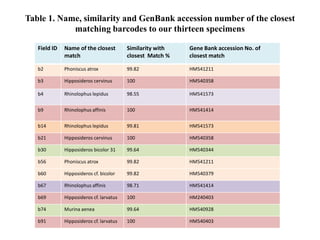
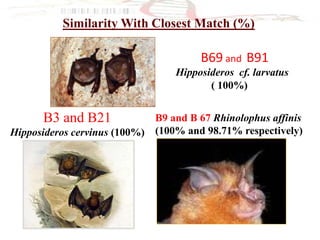
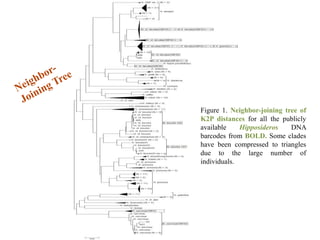
The document discusses the application of DNA barcoding, introduced by Paul Herbert in 2003, to identify bat species in Peninsular Malaysia, highlighting its utility in recognizing cryptic taxa. A study identified nine bat species from samples collected, confirming high biodiversity, with 44% of the species classified as 'dark taxa'. The integration of DNA barcoding in biodiversity inventories enhances species recognition at a low cost, facilitating conservation efforts.