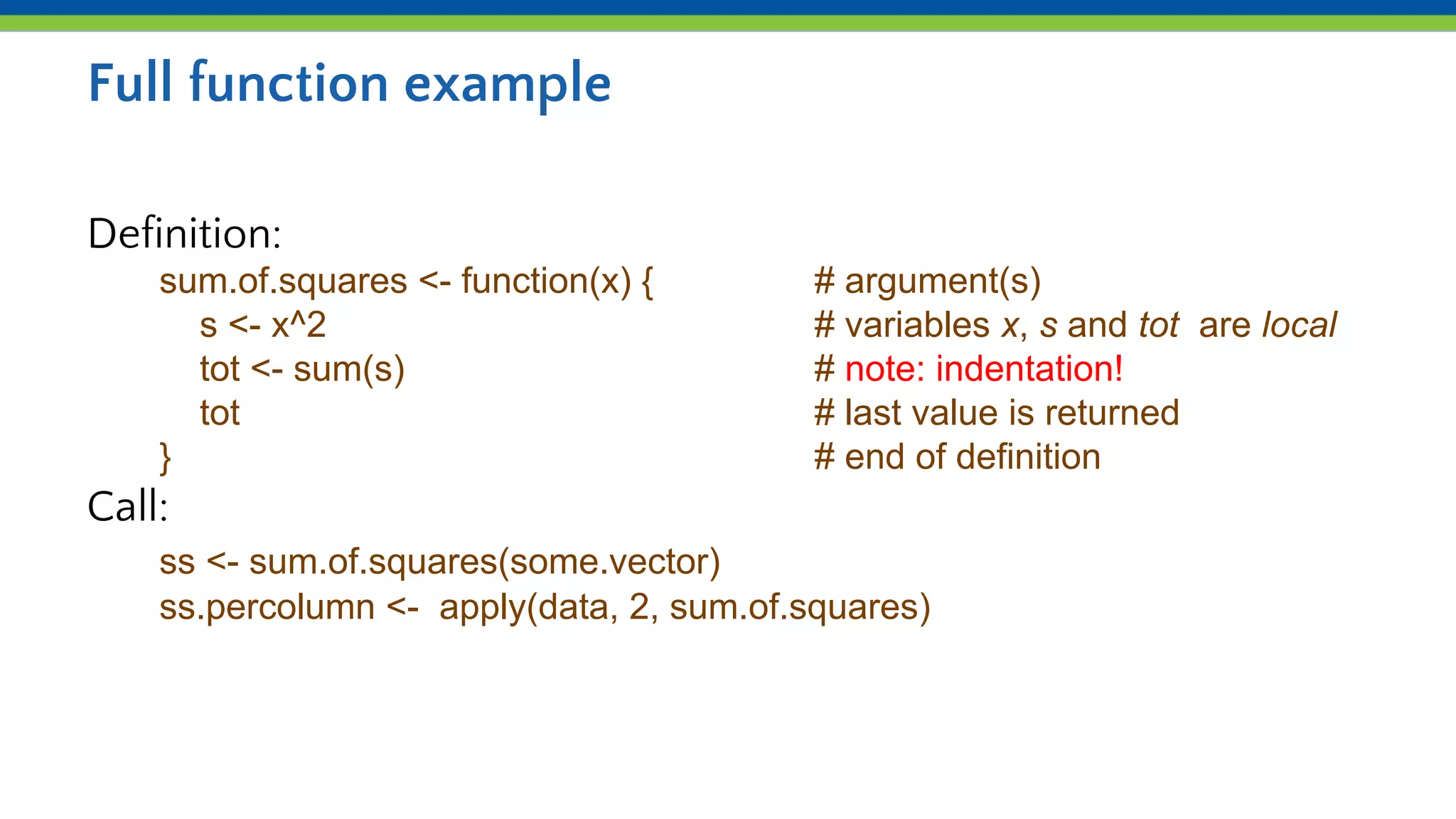
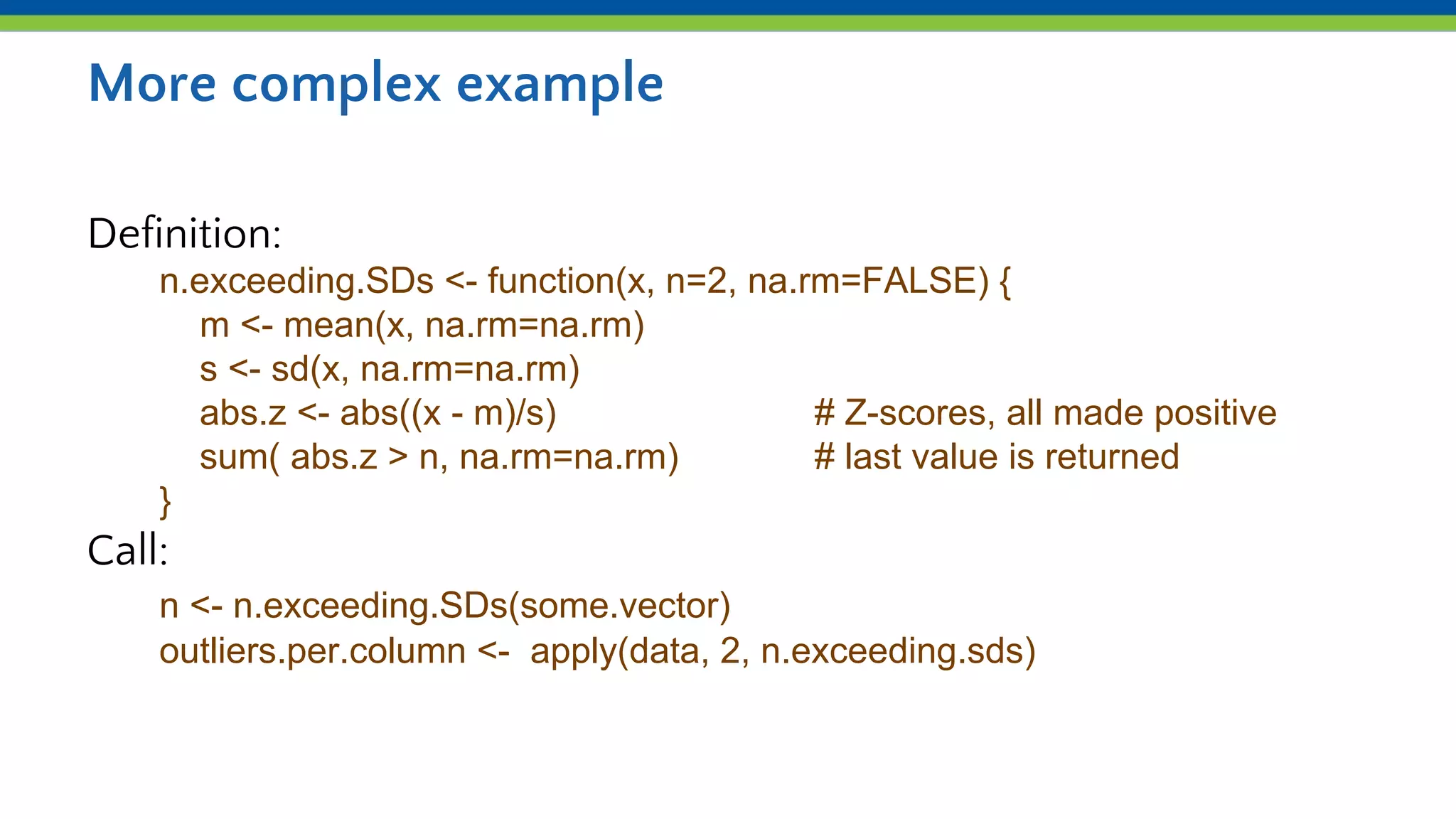
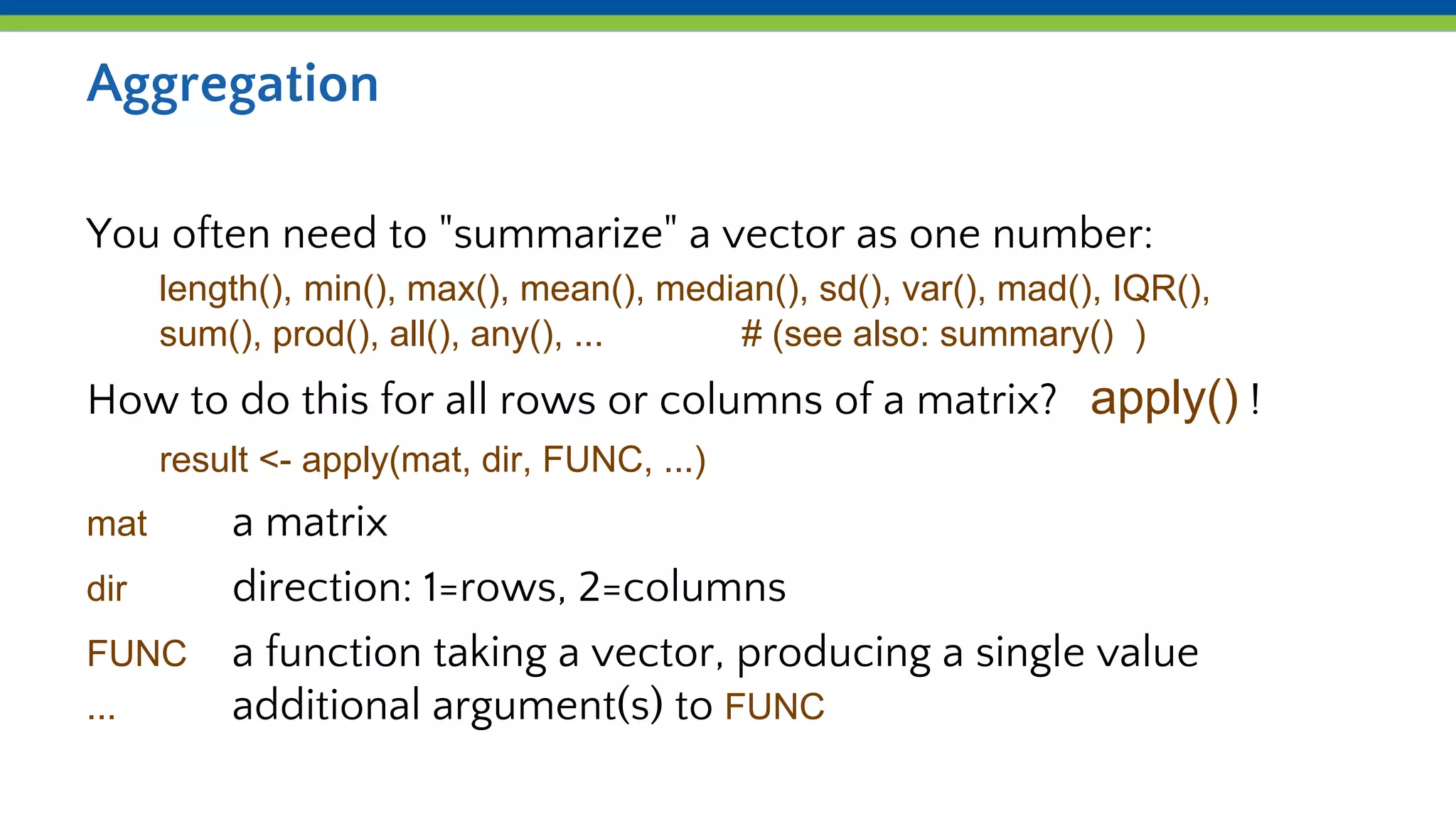
Iteration and functions allow repeating processes over sets of values in R. Functions define reusable recipes that automate actions, taking inputs as arguments and returning outputs. apply() applies a function over the rows or columns of a matrix or dataframe, allowing aggregation like summing or averaging. lapply() and sapply() similarly apply a function over the elements of a list. Custom functions can be defined and then used with apply() to iterate complex processes like counting values exceeding a number of standard deviations from the mean.
![What is iteration
Repeating a process to a set of values
Often automatic:
c <- a + b # Works for vectors and matrices
Reminder: value-recycling
matrix(1:4, nrow=2, byrow=TRUE) + c(70,90) →
[,1] [,2]
[1,] 71 72
[2,] 93 94 # (the recycling is column-wise!)](https://image.slidesharecdn.com/day4a-iterationandfunctions-171210231748/75/Day-4a-iteration-and-functions-pptx-2-2048.jpg)
![apply() cont’d
> m <- matrix(1:6, nrow=2)
> m
[,1] [,2] [,3]
[1,] 1 3 5 # filled column-wise by default
[2,] 2 4 6
> apply(m, 1, sum) # row-wise sum
[1] 9 12
> apply(m, 2, prod, na.rm=TRUE) # column-wise product, ignoring NA’s
[1] 2 12 30
And also:
lapply(lst, FUNC) # iterate over list-contents
tapply(), sapply(), mapply(), …](https://image.slidesharecdn.com/day4a-iterationandfunctions-171210231748/75/Day-4a-iteration-and-functions-pptx-4-2048.jpg)
![lapply() and sapply()
Works on lists, rather than matrices
> lst <- list(a=runif(7), b=runif(2), <etc>)
$a
[1] 0.971 0.380 0.287 0.787 0.721 0.938 0.364
$b
[1] 0.609 0.658
<etc>
> lapply(lst, mean)
$a
[1] 0.635
$b
[1] 0.633
<etc>](https://image.slidesharecdn.com/day4a-iterationandfunctions-171210231748/75/Day-4a-iteration-and-functions-pptx-5-2048.jpg)