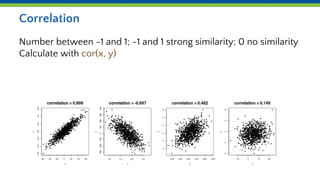
This document introduces various statistical functions in R including descriptive statistics like mean, median, and standard deviation. It covers distribution functions like the normal distribution and functions for generating random values. Hypothesis tests like the t-test are discussed along with ANOVA and linear models. Quantile functions and plotting are also introduced for understanding data distributions and removing outliers.


![Randomization of existing values
> sample(1:10)
[1] 8 3 5 7 4 9 6 1 10 2
> sample(1:10, size=4)
[1] 1 9 7 6
> sample(c(TRUE, FALSE), 6, replace=TRUE)
[1] FALSE TRUE FALSE TRUE FALSE FALSE
> sample(c("A", "C", "G", "T"), 16, replace=TRUE)
[1] "T" "T" "A" "T" "C" "C" "A" "G" "A" "G" "C" "A" "A" "T" "G" "C"](https://image.slidesharecdn.com/day5b-statisticalfunctions-171210233235/85/Day-5b-statistical-functions-pptx-6-320.jpg)

![Trimming
> quantile(x, probs=c(0.05, 0.95))
5% 95%
-1.404072 1.879870
> limits <- quantile(x, probs=c(0.05, 0.95))
> x <- x [ x > limits[1] & x < limits[2] ]
← these are just the names ...](https://image.slidesharecdn.com/day5b-statisticalfunctions-171210233235/85/Day-5b-statistical-functions-pptx-8-320.jpg)



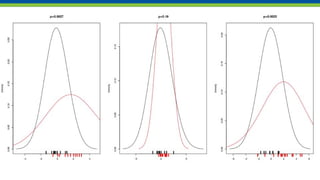
![tests return a list(), but print something else
t.test(x,y) →
Welch Two Sample t-test
data: x and y
t = -2.8096, df = 15.245, p-value = 0.01304
alternative hypothesis: true difference in means
is not equal to 0
95 percent confidence interval:
-2.0611160 -0.2843106
sample estimates:
mean of x mean of y
-0.08099273 1.09172057
> str(t)
List of 9
$ statistic : Named num -2.81
$ parameter : Named num 15.2
$ p.value : num 0.013
$ conf.int : atomic [1:2] -2.061 -0.284
$ estimate : Named num [1:2] -0.081 1.092
$ null.value : Named num 0
$ alternative: chr "two.sided"
$ method : chr "Welch Two Sample t-test"
$ data.name : chr "x and y"](https://image.slidesharecdn.com/day5b-statisticalfunctions-171210233235/85/Day-5b-statistical-functions-pptx-15-320.jpg)