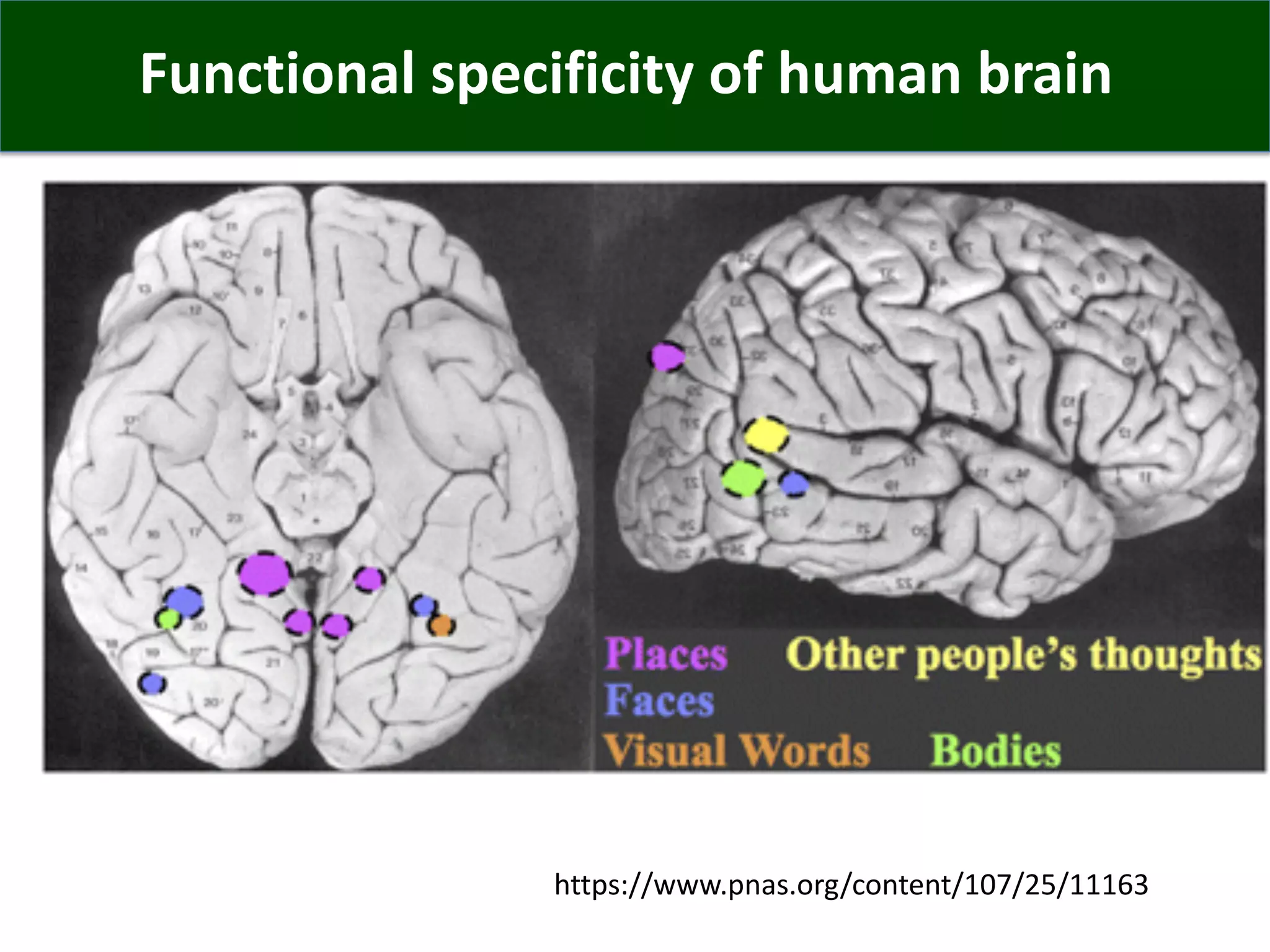
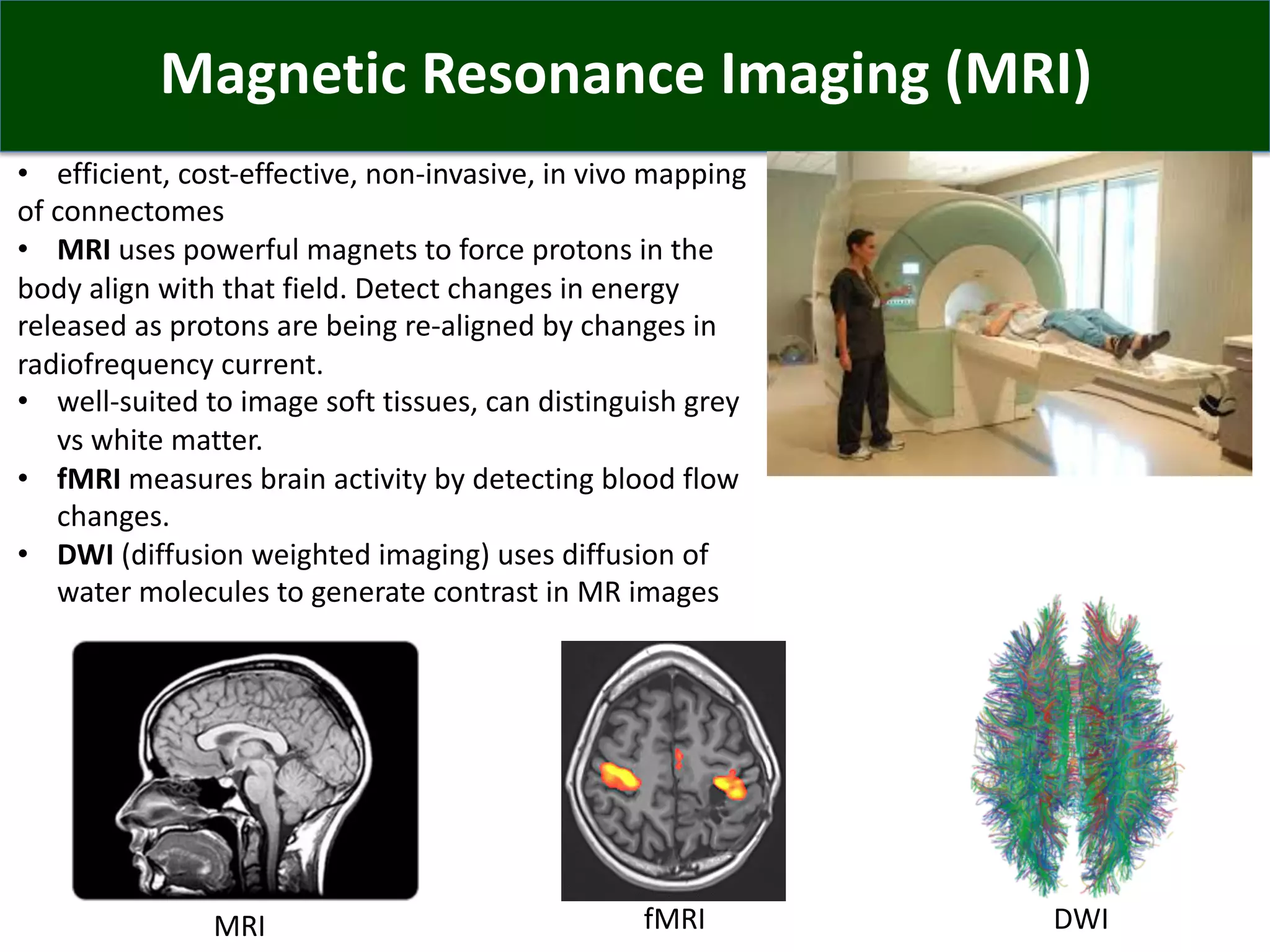
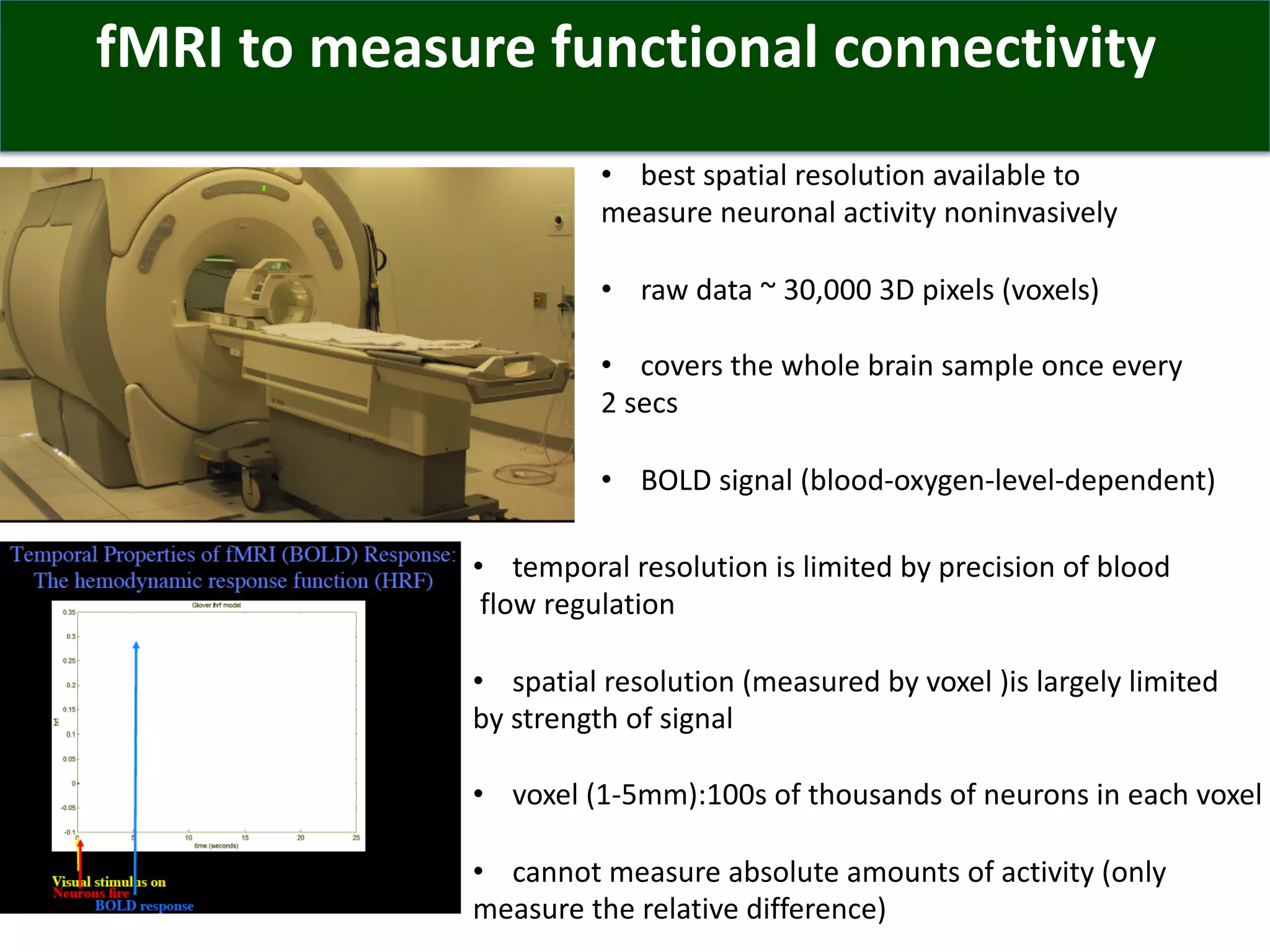
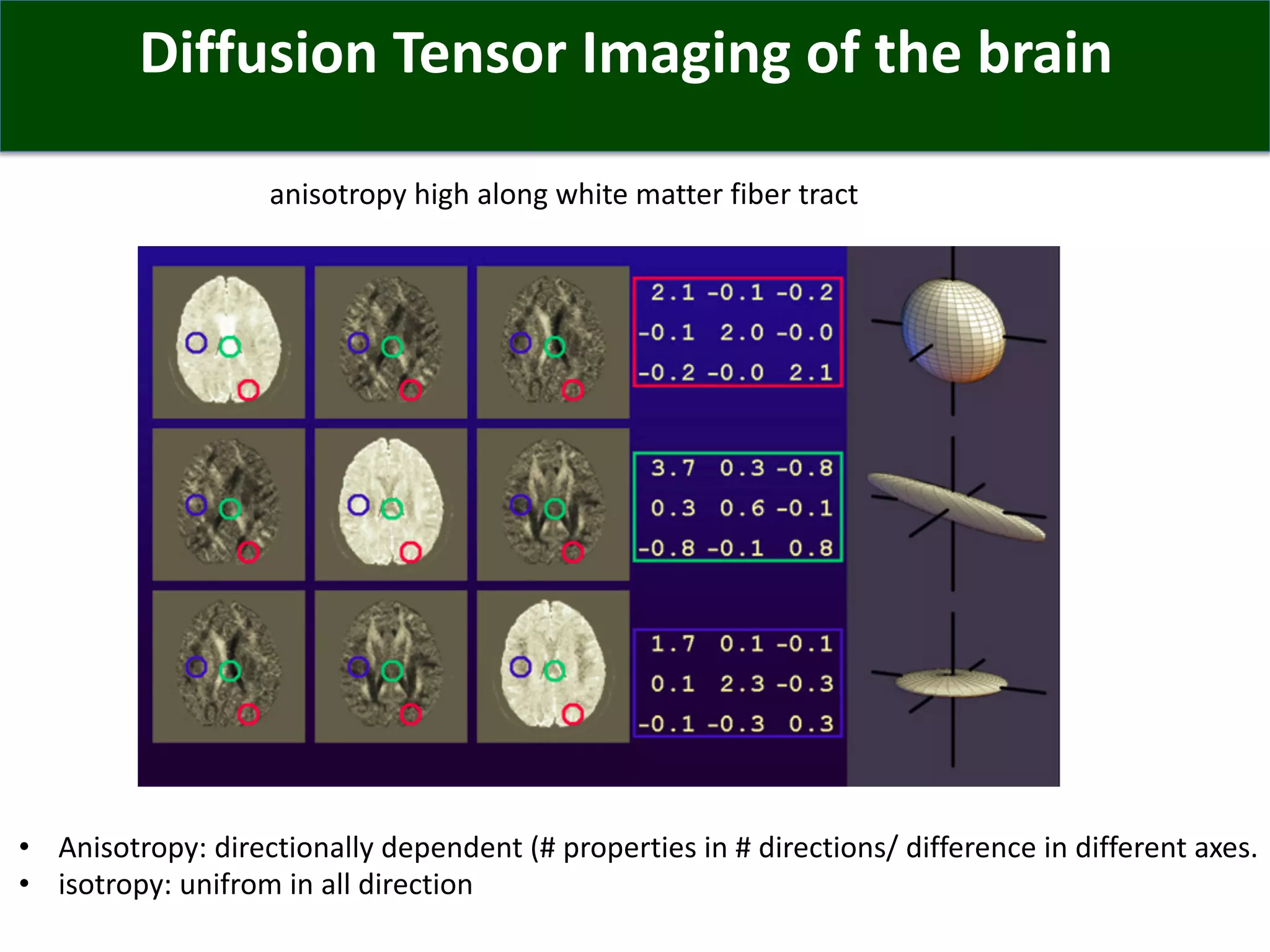
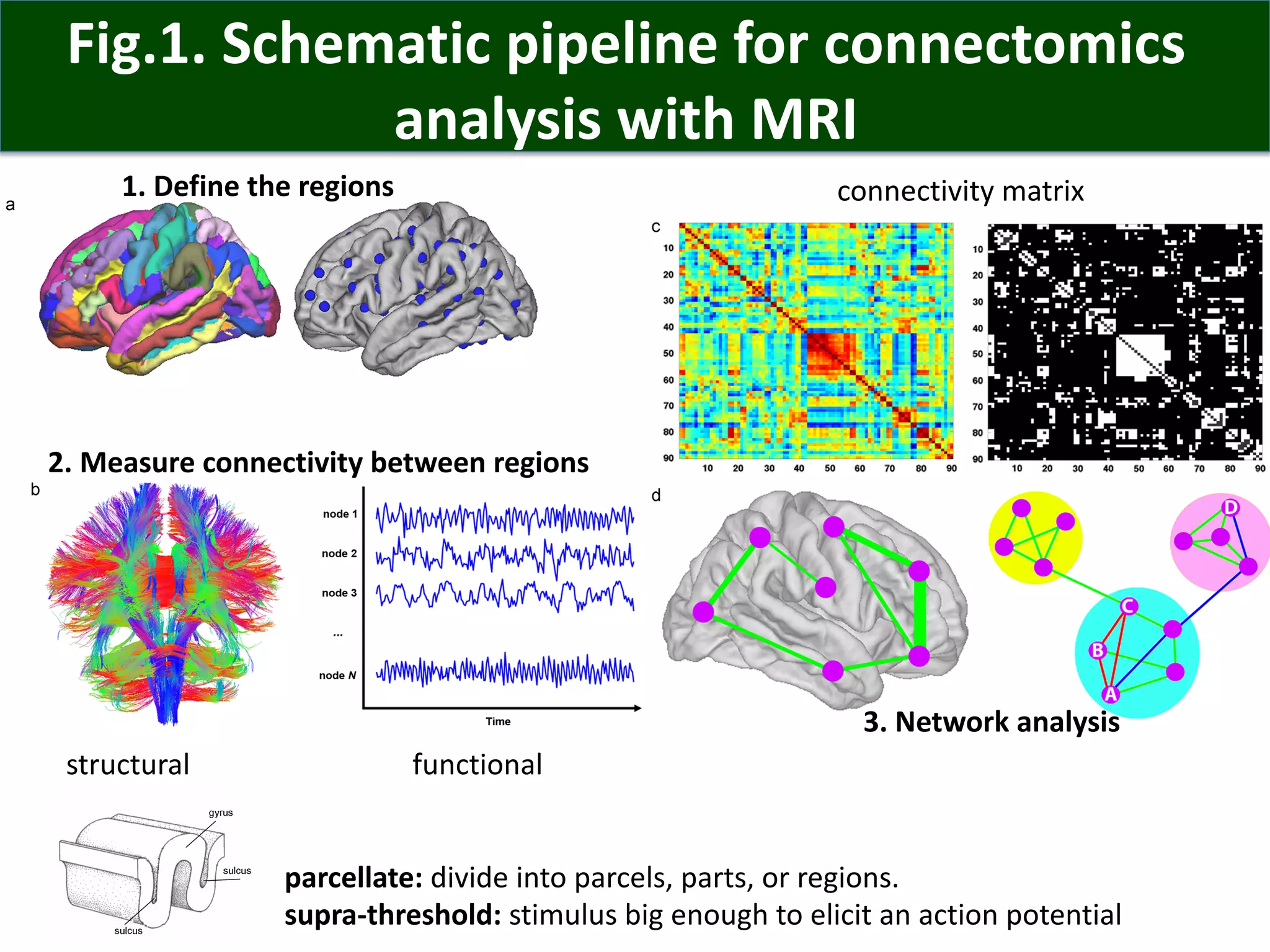
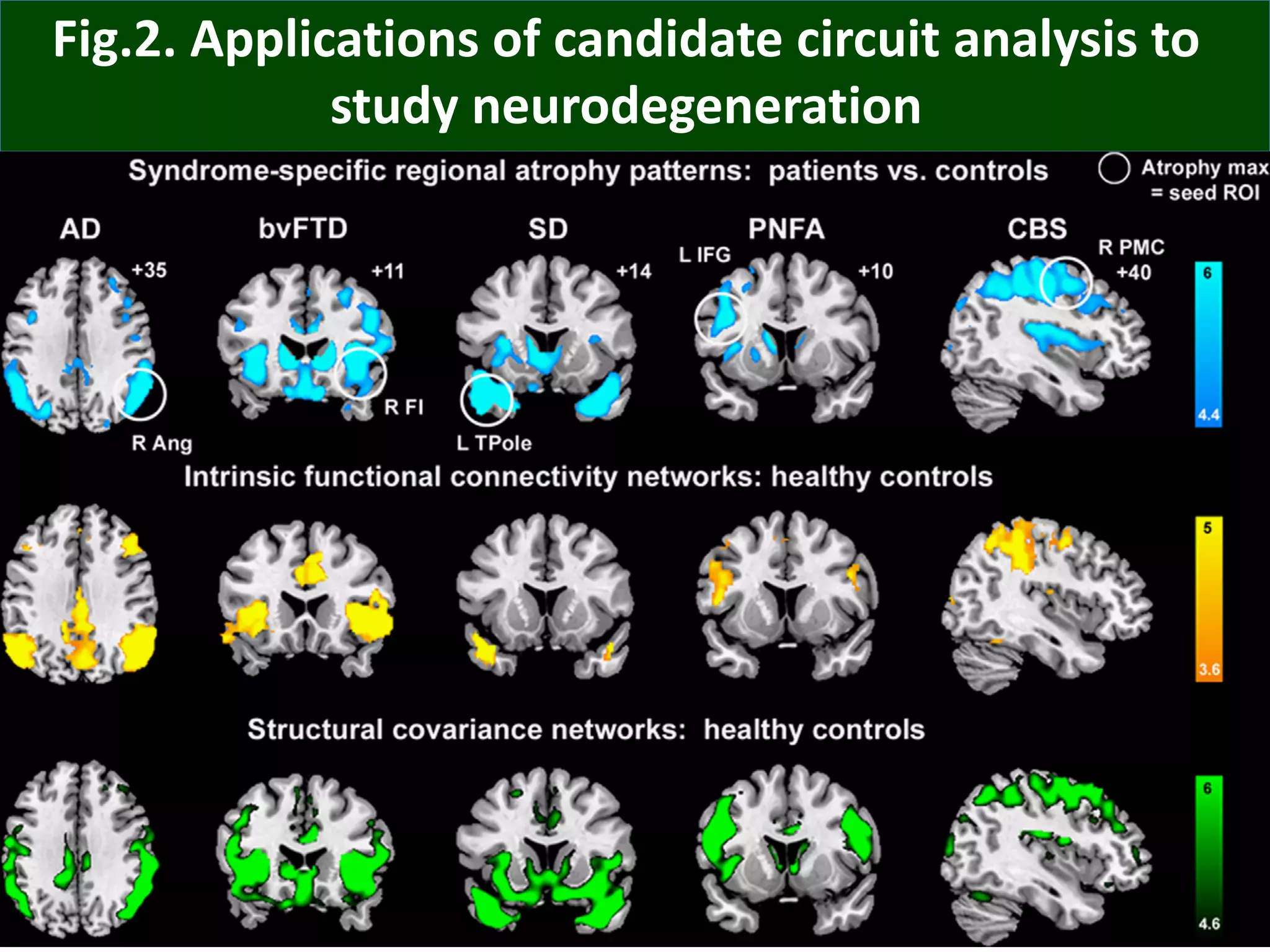
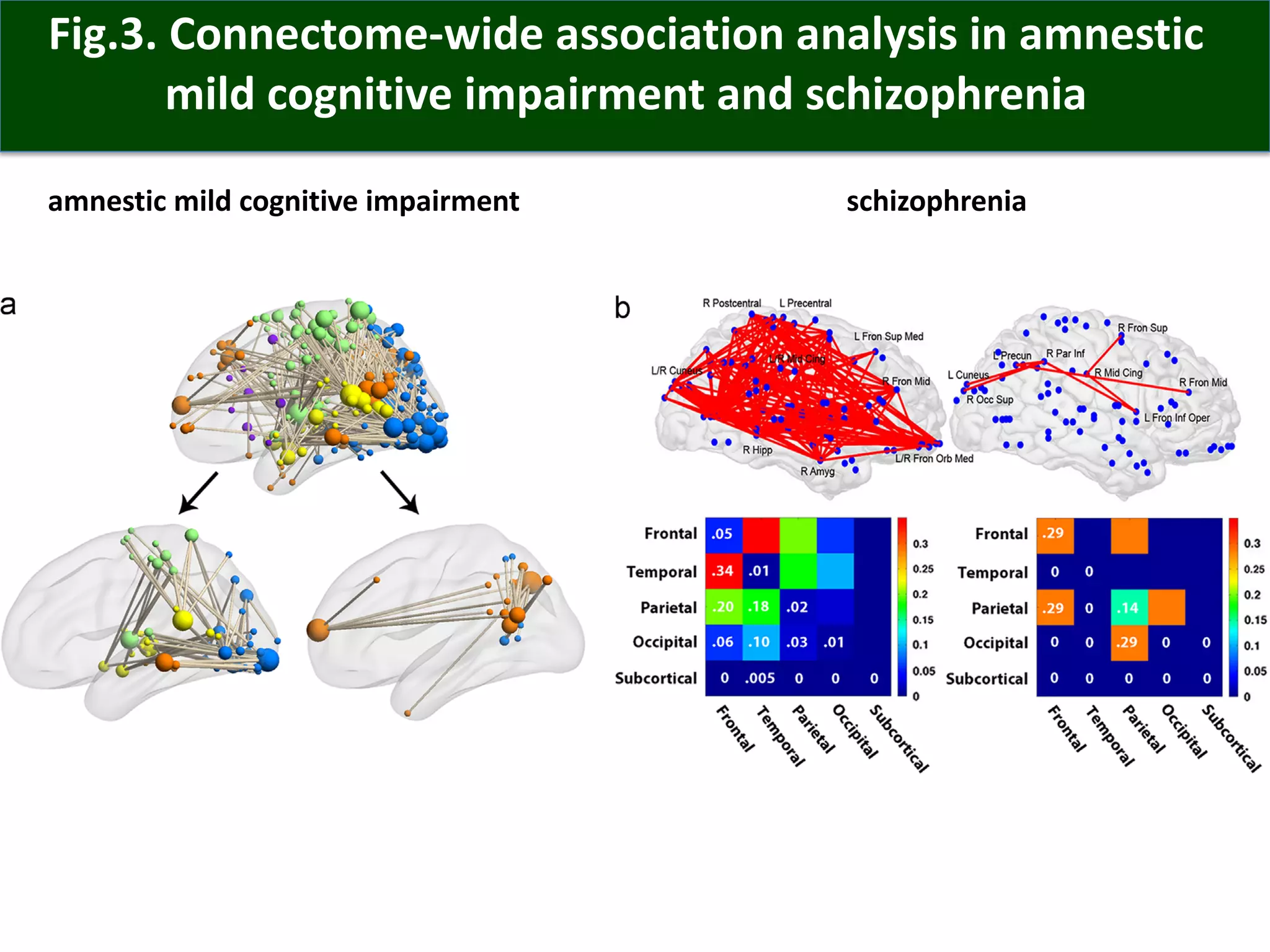
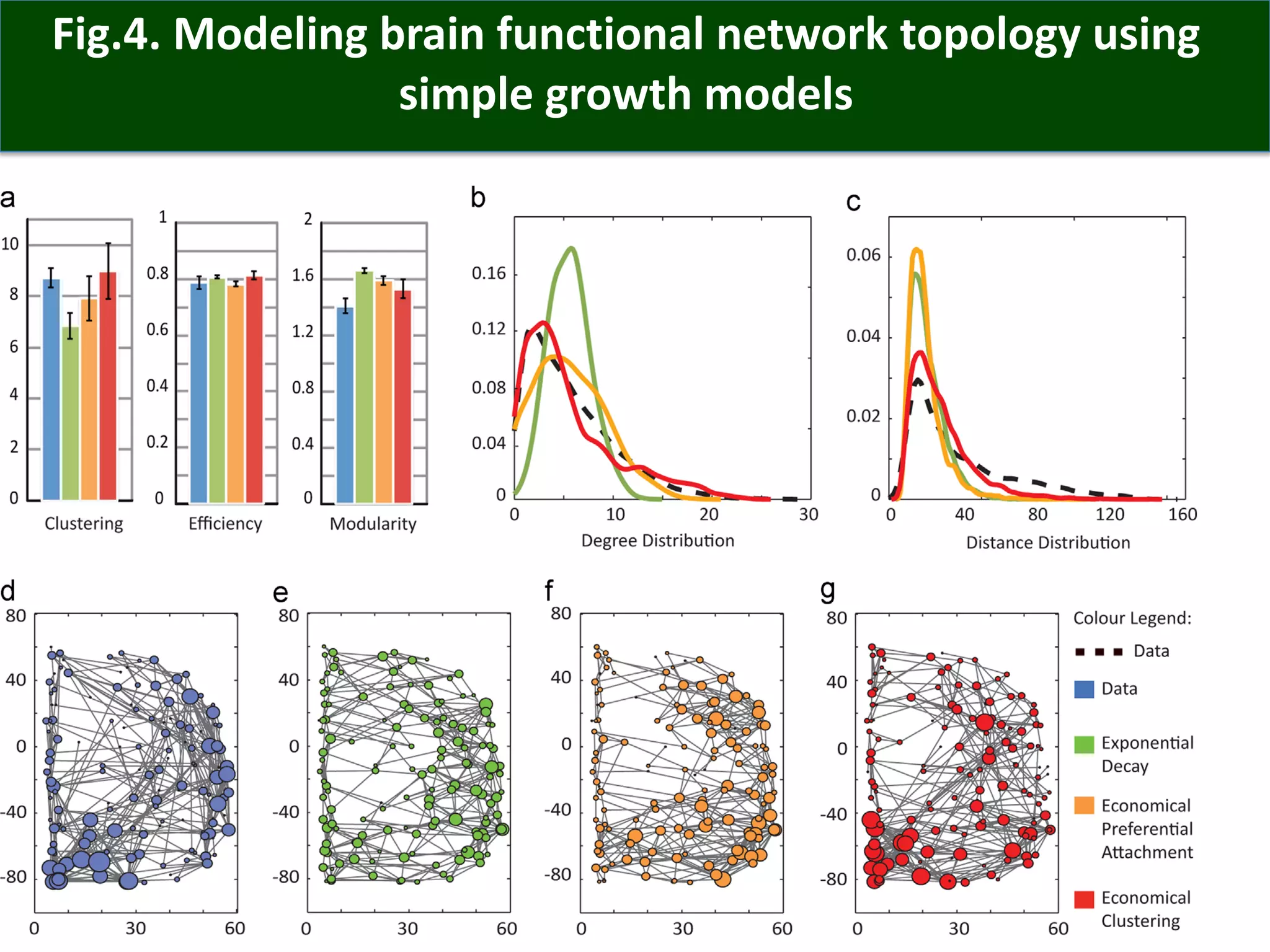
Connectomics is a new paradigm for understanding brain diseases using comprehensive maps of neural connections in the brain. Magnetic resonance imaging (MRI) techniques like functional MRI and diffusion MRI allow non-invasive mapping of functional and anatomical connectivity networks in the living human brain. There are three main approaches to analyzing connectome data: candidate circuit analysis of specific hypothesized networks, connectome-wide analysis of effects across the entire connectivity matrix, and topological analysis of network organization. Connectomics research aims to uncover mechanisms of neurological disorders and enable more targeted clinical applications.