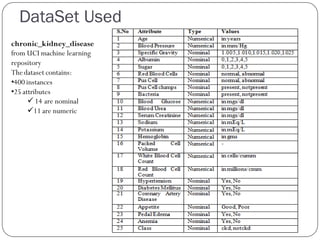
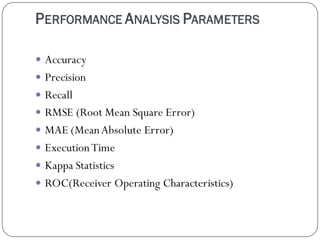
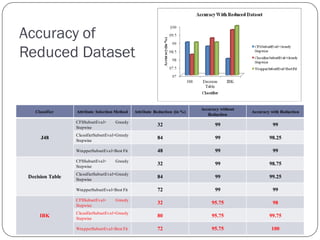
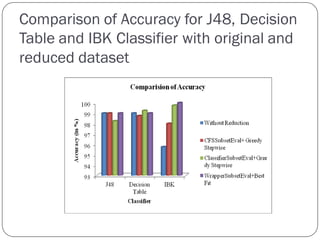
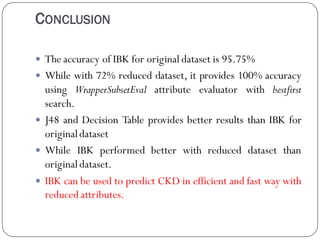
This document discusses using data mining classifiers and attribute reduction techniques to predict chronic kidney disease (CKD) more accurately and efficiently. It first provides background on CKD and the need for early detection. It then discusses data mining, classification algorithms, attribute selection filters and wrappers. The document analyzes several studies that predicted CKD using techniques like decision trees, SVM and Naive Bayes. It describes the dataset used from the UCI repository and evaluation metrics. The results section compares J48, Decision Tree and IBK classifiers with and without attribute reduction using CfsSubsetEval, ClassifierSubsetEval and WrapperSubsetEval. Attribute reduction improved accuracy, especially for IBK which achieved 100% accuracy with 72% fewer attributes.

![Introduction
As the past records show, the number of deaths in
India due to chronic kidney disease (CKD) were 5.21
million in 2008 and this number can be further
raised to 7.63 million by 2020 [4] .
There is need of detection of the chronic kidney
disease at early stage before getting it worse.
To reduce mortality rate, an efficient technique is required
to predict and classify it.](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-3-320.jpg)
![Data Mining & Classification
Data mining refers to extracting meaningful
information from hidden patterns of dataset [2].
The data mining techniques are very useful in health
informatics [16, 17].
Data mining classification techniques play a vital role
in classifying various diseases from symptoms and
various medical tests.](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-7-320.jpg)





![Literature Review
Researcher Year Classifier Accuracy Remarks
K.R. Lakshmi [6] 2014 ANN 93.8521% Performed better than
Decision Tree and Logical
regressionclassifiers
Naganna Chetty
[7]
2015 NaïveBayes,
SMO,IBK
99%,98.25%,
100%
Attribute Reduction using
Wrapper Method
S.Vijayarani [8] 2015 SVM 76.32%. 584 instances and six
attributes
L.Jerlin Rubini
[9]
2015 Multilayer
Preceptor
99.75% Performed better than radial
basis function network, logistic
regression
Uma N Dulhare
[10]
2016 NaïveBayes 97.5% Attribute Reduction using
OneR
HuseyinPolat
[11]
2017 SVM 98.5%. Attribute Reduction
WalaA. [12] 2017 Decisiontree 99% Missing Values are replaced
withmean](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-13-320.jpg)


![References
[1] L. Jena, and N. Ku. Kamila, "Distributed data mining classification algorithms for prediction of chronic-
kidney-disease," International Journal of Emerging Research in Management &Technology, vol-4, Issue-
11, pp: 110-118, November 2015.
[2] K. Chandel, V. Kunwar, S. Sabitha, T. Choudhury, and S. Mukherjee, “A comparative study on thyroid
disease detection using K-nearest neighbor and Naive Bayes classification techniques, CSI transactions on
ICT, 4(2-4), pp: 313-319, 2016.
[3] Sudhir B. Jagtap, "Census data mining and data analysis using WEKA," arXiv preprint arXiv:1310.4647,
2013.
[4] S.Dilli Arasu, R.Thirumalaiselvi, “Review of Chronic Kidney Disease based on Data Mining Techniques,”
International Journal ofApplied Engineering Research, vol-12, pp: 13498-13505, 2017.
[5] S. Zeynu, Shruti Patil, “Survey on Prediction of Chronic Kidney Disease Using Data Mining Classification
Techniques and Feature Selection,” International Journal of Pure and Applied Mathematics, vol-118, No.
8,pp:149-156, 2018.
[6] K. R. Lakshmi, Y. Nagesh, and M. Veera Krishna, "Performance comparison of three data mining techniques
for predicting kidney dialysis survivability," International Journal of Advances in Engineering &
Technology, vol. 7, pp: 242-254, 2014.
[7] N. Chetty, Kunwar Singh Vaisla, and Sithu D. Sudarsan, “Role of attributes selection in classification of
Chronic Kidney Disease patients,” Computing, Communication and Security (ICCCS), International
Conference on. IEEE, 2015.](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-22-320.jpg)
![References
[8] S. Vijayarani, and S. Dhayanand, "Data mining classification algorithms for kidney disease
prediction,"International Journal on Cybernetics and Informatics (IJCI) , 2015.
[9] L. Jerlin Rubini and Dr. P. Eswaran, “Generating comparative analysis of early stage prediction of Chronic
Kidney Disease,” International Journal of Modern Engineering Research (IJMER), Volume 5, Issue 7, pp
49-55, July2015.
[10] Uma N. Dulhare, and Mohammad Ayesha, “Extraction of action rules for chronic kidney disease using
Naïve bayes classifier,” Computational Intelligence and Computing Research (ICCIC), IEEE International
Conference on IEEE, 2016.
[11] H. Polat, Homay Danaei Mehr, and Aydin Cetin, “Diagnosis of chronic kidney disease based on support
vector machine by feature selection methods,” Journal of medical systems, Feb 2017.
[12] W. Abedalkhader, and Noora Abdulrahman, “Missing Data Classification Of Chronic Kidney Disease,”
International Journal of Data Mining & Knowledge Management Process (IJDKP) Vol.7, No.5/6,
November 2017.
[13] Abeer Y. Al-Hyari, “Chronic Kidney Disease Prediction System UsingClassifying Data Mining Techniques,”
Library of university of Jordan, 2012.
[14 Jiliang Tang, Salem Alelyani, and Huan Liu, “Feature selection for classification: A review,” Data
classification:Algorithms and applications, 2014.](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-23-320.jpg)
![References
[15] Geoffrey Holmes, Andrew Donkin, and Ian H. Witten, “Weka: A machine learning workbench,”
Intelligent Information Systems, 1994. Proceedings of the 1994 Second Australian and New Zealand
Conference on. IEEE, 1994.
[16] Mary K. Obenshain, “Application of data mining techniques to healthcare data,” Infection Control &
Hospital Epidemiology25.8, pp: 690-695, 2004.
[17] Cheng, Li-Chen, Ya-Han Hu, and Shr-Han Chiou, “Applying the Temporal Abstraction Technique to the
Predictionof Chronic Kidney Disease Progression,” Journal of medical systems 41, April 2017.
[18] Neeraj Bhargava, Girja Sharma, Ritu Bhargava, and Manish Mathuria, “Decision tree analysis on J48
algorithm for data mining,” Proceedings of International Journal of Advanced Research in Computer
Scienceand Software Engineering, Vol. 3, pp:1114-1119, June 2013.
[19] Hongjun Lu, and Hongyan Liu, “Decision tables: Scalable classification exploring RDBMS
capabilities,”Proceedings of the 26th International Conference onVery Large Data Bases,VLDB'00. 2000.](https://image.slidesharecdn.com/ckd-ppt-190304031435/85/Chronic-Kidney-Disease-Prediction-24-320.jpg)