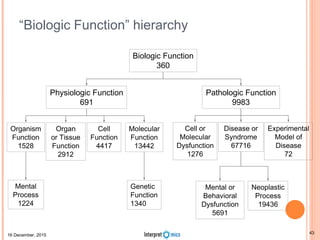
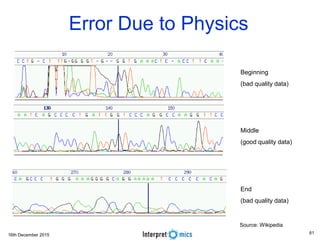
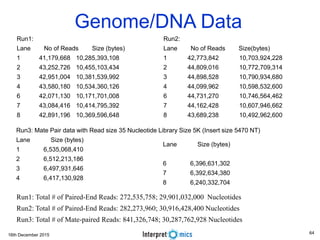
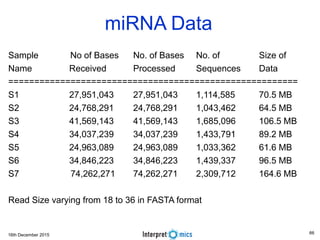
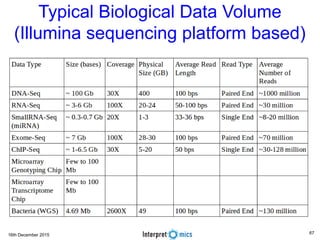
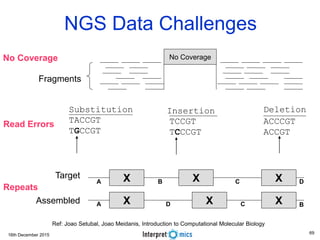
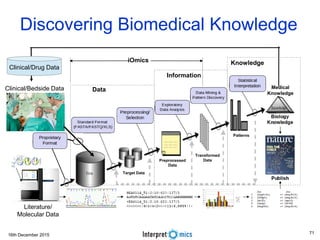
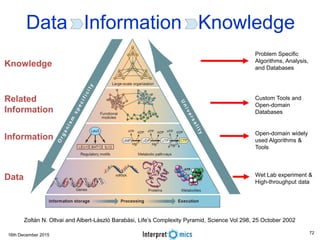
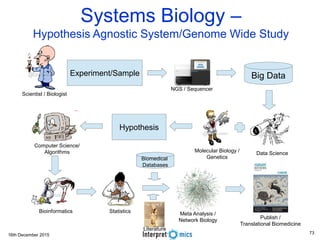
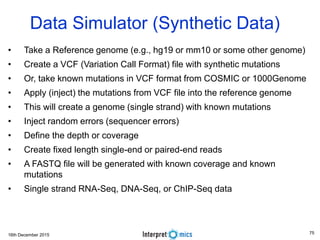
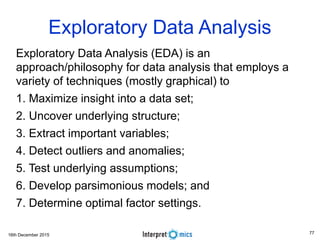
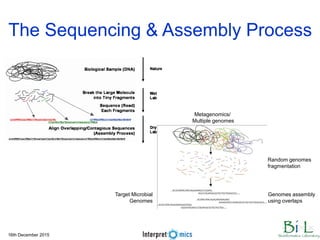
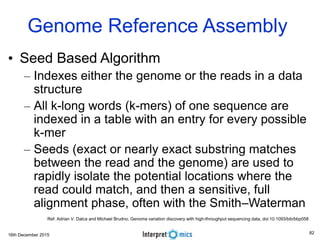
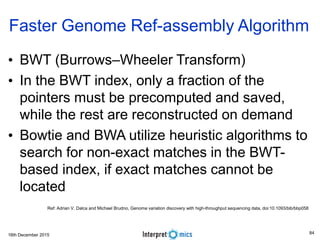
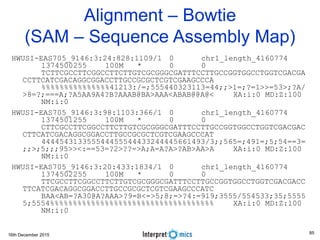
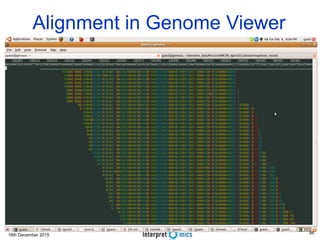
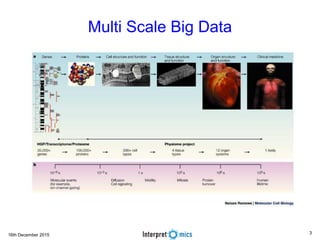
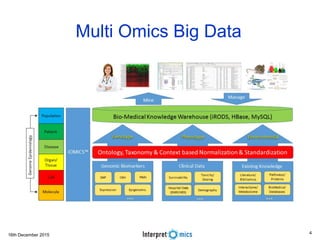
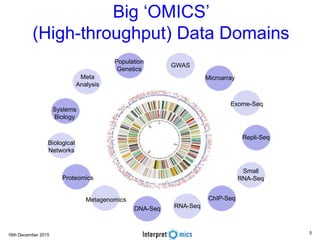
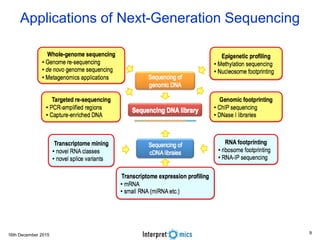
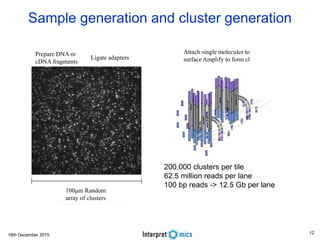
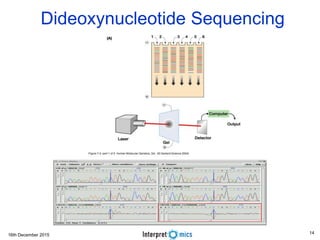
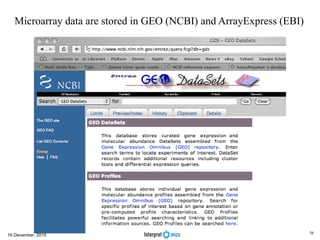
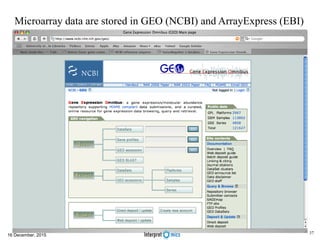
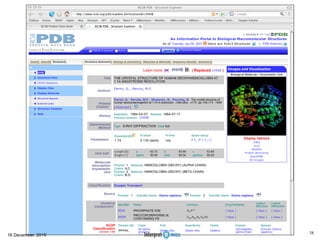
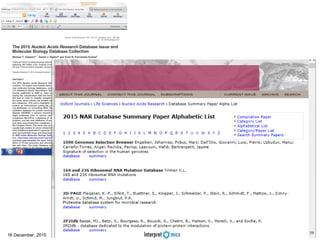
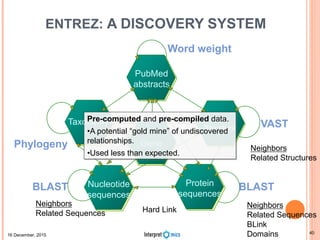
This document summarizes a presentation on genomics and big data in precision medicine. It discusses how next generation sequencing is generating massive amounts of multi-omics data from the genome, epigenome, transcriptome, proteome and metagenome. It describes some of the algorithms and databases used to analyze this big genomic and biological data, including de Bruijn graph algorithms and databases like NCBI, OMIM, and PANTHER. It also discusses some of the challenges in analyzing such large and complex biological data using computational methods.

![16th December 2015
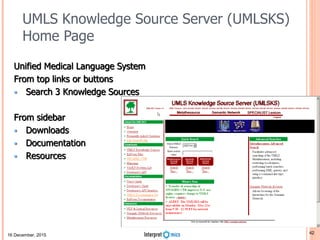
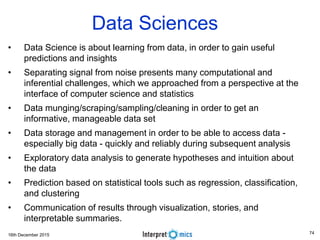
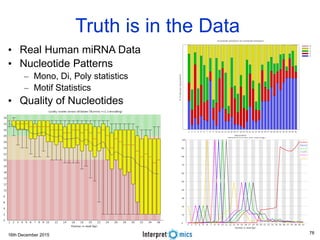
Dimensions of Big Data
The 7 Vs of Genomic Big Data
• Volume is defined in terms of the physical volume of the data that need to be online, like giga-byte (10
9 ), tera-byte (10 12 ), peta-byte (10 15 ) or exa-byte (10 18 ) or even beyond.
• Velocity is about the data-retrieval time or the time taken to service a request. Velocity is
also measured through the rate of change of the data volume.
• Variety relates to heterogeneous types of data like text, structured, unstructured, video, audio
etcetera.
• Veracity is another dimension to measure data reliability - the ability of an organization to trust the data
and be able to confidently use it to make crucial decisions.
• Vexing covers the effectiveness of the algorithm. The algorithm needs to be designed to ensure that
data processing time is close to linear and the algorithm does not have any bias; irrespective of the
volume of the data, the algorithm is able to process the data in reasonable time.
• Variability is the scale of data. Data in biology is multi-scale, ranging from sub-atomic ions at
picometers, macro-molecules, cells, tissues and finally to a population [9] at thousands of kilometers.
• Value is the final actionable insight or the functional knowledge. The same
mutation in a gene may have a different effect depending on the population or the
environmental factors.](https://image.slidesharecdn.com/bda2015-tutorial-part2-datadatabases-160311113748/85/Bda2015-tutorial-part2-data-amp-databases-7-320.jpg)








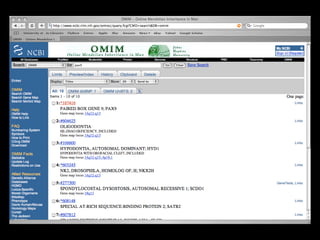
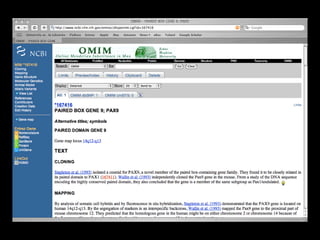
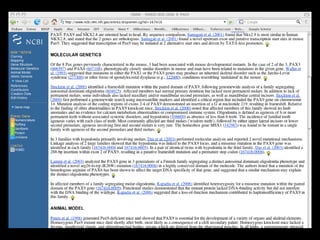
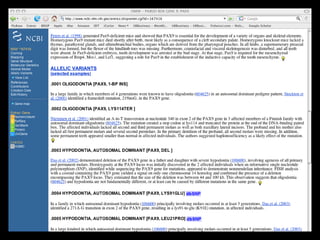
![PRECISE RESULTS
MLH1[Gene Name] AND Human[Organism]](https://image.slidesharecdn.com/bda2015-tutorial-part2-datadatabases-160311113748/85/Bda2015-tutorial-part2-data-amp-databases-41-320.jpg)