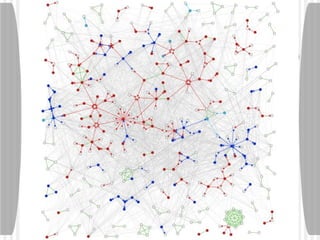
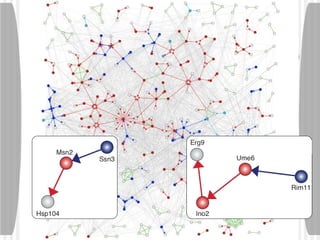
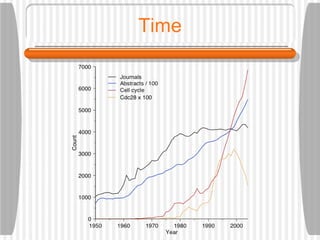
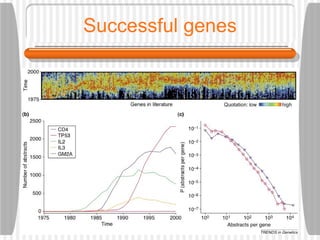
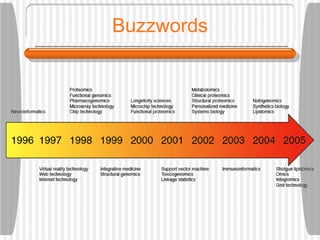
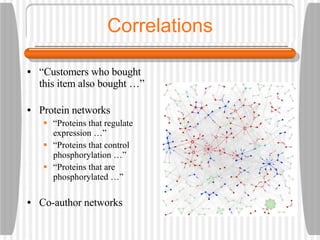
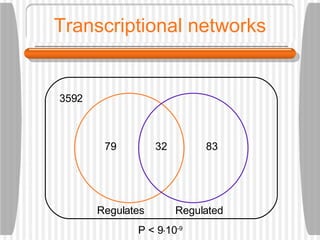
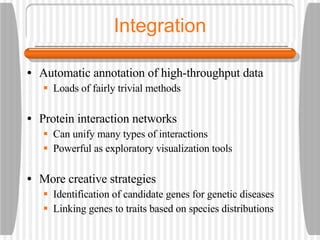
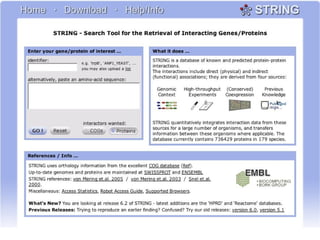
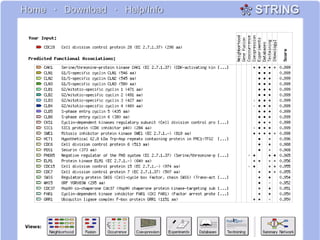
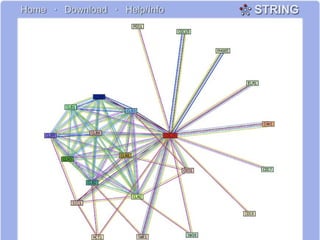
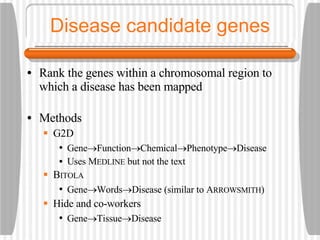
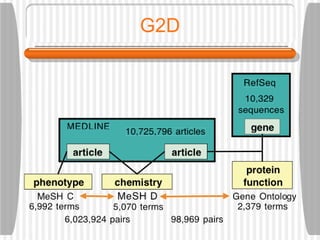
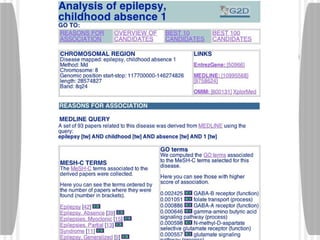
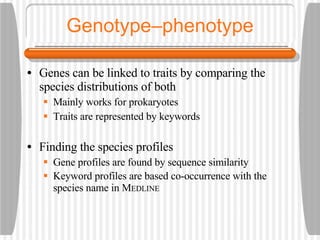
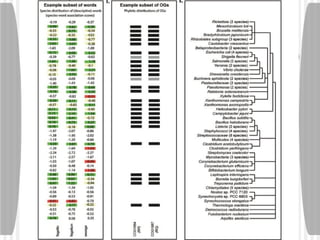
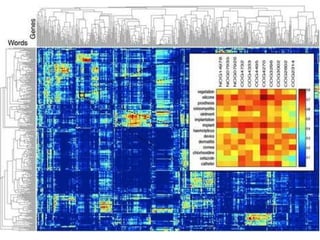
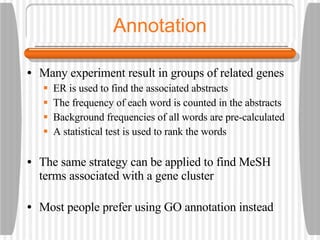
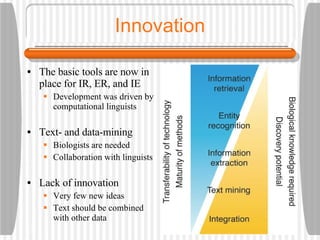
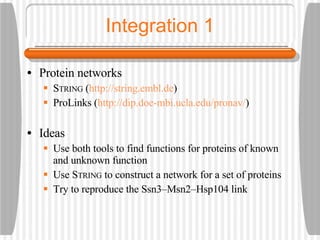
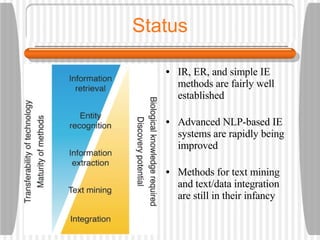
This document discusses various methodologies for extracting information from biological literature, including information retrieval, entity recognition, information extraction, and text/data mining. It provides an overview of different approaches like using co-occurrence, natural language processing, and machine learning methods. It also discusses challenges like integrating text with other data types and dealing with issues like ambiguity. Examples of existing text mining tools and their potential applications are also described.












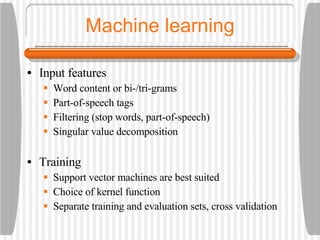
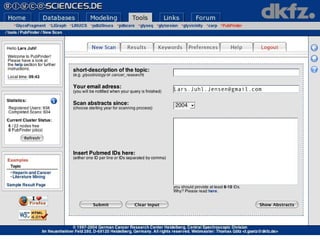







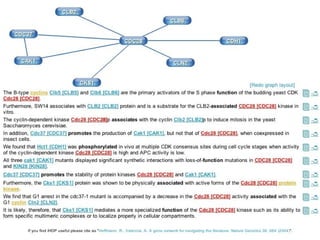



![Semantic labeling Gene and protein names Cue words for entity recognition Cue words for relation extraction Named entity chunking A CASS grammar recognizes noun chunks related to gene expression: [ nxgene The GAL4 gene ] Relation chunking Our CASS grammar also extracts relations between entities: [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]](https://image.slidesharecdn.com/jen06talk1-1217969253936187-8/85/Biomedical-literature-mining-40-320.jpg)
![[ expression_repression_active Btk regulates the IL-2 gene ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors ] [ expression_activation_passive [ expression IL-13 expression] induced by IL-2 + IL-18 ] [ expression_activation_passive [ expression IL-13 expression ] induced by IL-2 + IL-18 ] [ expression_repression_active Btk regulates the IL-2 gene ]](https://image.slidesharecdn.com/jen06talk1-1217969253936187-8/85/Biomedical-literature-mining-41-320.jpg)