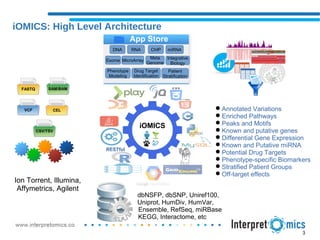
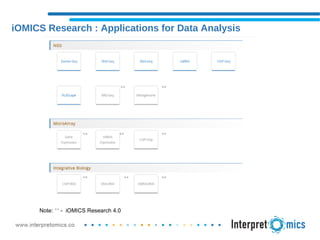
iOMICS Research is a unified software for multi-omics data analysis and interpretation. It provides intuitive analysis and dynamic visualization of DNA, RNA, ChIP, miRNA, and other omics data types. The software integrates proprietary and third-party databases for annotation and runs on both cloud and on-premise systems. It offers flexible, customizable workflows to efficiently process large sample sizes from raw data through analysis to results visualization.