Analytical Validation of the Oncomine™ Comprehensive Assay v3 with FFPE and Cell Line Tumor Specimens in a CAP-Accredited and CLIA-Certified Clinical Laboratory
•
1 like•1,588 views
The document summarizes an analytical validation of the Oncomine Comprehensive Assay v3 (OCAv3) targeted next-generation sequencing panel performed in a CLIA-certified laboratory. The validation assessed analytical sensitivity, specificity, accuracy, and precision using formalin-fixed paraffin-embedded tumor samples and cell lines. Results showed the assay met performance thresholds of 90% or higher for detecting single nucleotide variants, insertions/deletions, copy number variants, and gene fusions across a wide range of variants. Over 2,500 clinical samples were subsequently sequenced with the assay maintaining a 95% success rate and average turnaround time of 10 days.
Report
Share
Report
Share
Download to read offline
Recommended
Increasing genome editing efficiency with optimized CRISPR-Cas enzymes

Genome editing by CRISPR systems has proven to be groundbreaking for basic biomedical research with significant implications for the treatment of human diseases. While the CRISPR-Cas9 and CRISPR-Cas12a (Cpf1) systems enable genome editing in a broad range of host species and cell types, both can exhibit poor editing efficiencies at specific target sites or in systems where delivery of CRISPR reagents is difficult. There are concerns about target specificity of the CRISPR-Cas9 system and, in many cases, typical remedies such as modified guide RNAs or mutant Cas9 proteins cause loss of genome editing efficiency. Many of these solutions for improving specificity were developed for delivery of the Cas9-gRNA complex via plasmid DNA vectors rather than delivery as ribonucleoproteins (RNPs). However, RNP delivery of CRISPR reagents is being increasingly used because of the risk of unwanted stimulation of the immune system by plasmid delivery.
In this webinar, Dr Vakulskas discusses improved Cas9 and Cas12a (Cpf1) nucleases that have been optimized to significantly increase editing efficiency in living cells. He also presents data showing that IDT’s latest high-fidelity Cas9, Alt-R HiFi S.p. Cas9 V3, increases on-target editing efficiency and dramatically reduces off-target editing.
Clinical Applications of Next Generation Sequencing

Clinical Applications of Next Generation Sequencing
Applications of Single Cell Analysis

Single cell analysis has exploded recently mainly due to the development of high-throughput technologies such as NGS. Single cell analysis is being pursued by researchers in many areas including developmental science, cancer, biomarker discovery and more. This presentation covers some of the recent applications from developed by QIAGEN customers.
Introduction to NGS Variant Calling Analysis (UEB-UAT Bioinformatics Course -...

Introduction to NGS Variant Calling Analysis (UEB-UAT Bioinformatics Course -...VHIR Vall d’Hebron Institut de Recerca
Course: Bioinformatics for Biomedical Research (2014).
Session: 2.3- Introduction to NGS Variant Calling Analysis.
Statistics and Bioinformatisc Unit (UEB) & High Technology Unit (UAT) from Vall d'Hebron Research Institute (www.vhir.org), Barcelona.An introduction to RNA-seq data analysis

AGRF in conjunction with EMBL Australia recently organised a workshop at Monash University Clayton. This workshop was targeted at beginners and biologists who are new to analysing Next-Gen Sequencing data. The workshop also aimed to provide users with a snapshot of bioinformatics and data analysis tips on how to begin to analyse project data. An introduction to RNA-seq data analysis was presented by AGRF Senior Bioinformatician Dr. Sonika Tyagi.
Presented: 1st August 2012
Recommended
Increasing genome editing efficiency with optimized CRISPR-Cas enzymes

Genome editing by CRISPR systems has proven to be groundbreaking for basic biomedical research with significant implications for the treatment of human diseases. While the CRISPR-Cas9 and CRISPR-Cas12a (Cpf1) systems enable genome editing in a broad range of host species and cell types, both can exhibit poor editing efficiencies at specific target sites or in systems where delivery of CRISPR reagents is difficult. There are concerns about target specificity of the CRISPR-Cas9 system and, in many cases, typical remedies such as modified guide RNAs or mutant Cas9 proteins cause loss of genome editing efficiency. Many of these solutions for improving specificity were developed for delivery of the Cas9-gRNA complex via plasmid DNA vectors rather than delivery as ribonucleoproteins (RNPs). However, RNP delivery of CRISPR reagents is being increasingly used because of the risk of unwanted stimulation of the immune system by plasmid delivery.
In this webinar, Dr Vakulskas discusses improved Cas9 and Cas12a (Cpf1) nucleases that have been optimized to significantly increase editing efficiency in living cells. He also presents data showing that IDT’s latest high-fidelity Cas9, Alt-R HiFi S.p. Cas9 V3, increases on-target editing efficiency and dramatically reduces off-target editing.
Clinical Applications of Next Generation Sequencing

Clinical Applications of Next Generation Sequencing
Applications of Single Cell Analysis

Single cell analysis has exploded recently mainly due to the development of high-throughput technologies such as NGS. Single cell analysis is being pursued by researchers in many areas including developmental science, cancer, biomarker discovery and more. This presentation covers some of the recent applications from developed by QIAGEN customers.
Introduction to NGS Variant Calling Analysis (UEB-UAT Bioinformatics Course -...

Introduction to NGS Variant Calling Analysis (UEB-UAT Bioinformatics Course -...VHIR Vall d’Hebron Institut de Recerca
Course: Bioinformatics for Biomedical Research (2014).
Session: 2.3- Introduction to NGS Variant Calling Analysis.
Statistics and Bioinformatisc Unit (UEB) & High Technology Unit (UAT) from Vall d'Hebron Research Institute (www.vhir.org), Barcelona.An introduction to RNA-seq data analysis

AGRF in conjunction with EMBL Australia recently organised a workshop at Monash University Clayton. This workshop was targeted at beginners and biologists who are new to analysing Next-Gen Sequencing data. The workshop also aimed to provide users with a snapshot of bioinformatics and data analysis tips on how to begin to analyse project data. An introduction to RNA-seq data analysis was presented by AGRF Senior Bioinformatician Dr. Sonika Tyagi.
Presented: 1st August 2012
Exome seuencing (steps, method, and applications)

Exome sequencing, steps, Next generation sequencing
Rna seq pipeline

RNA Sequence data analysis,Transcriptome sequencing, Sequencing steady state RNA in a sample is known as RNA-Seq. It is free of limitations such as prior knowledge about the organism is not required.
RNA-Seq is useful to unravel inaccessible complexities of transcriptomics such as finding novel transcripts and isoforms.
Data set produced is large and complex; interpretation is not straight forward.
Pcr troubleshooting presentation

Presentation is about the pcr troubles which lead to false results..
Molecular Diagnostics.pptx

Molecular diagnostics is widely used for diagnosing genetic diseases and also the pathogenic diseases by using molecular tools.
Overcoming the challenges of designing efficient and specific CRISPR gRNAs

Overcoming the challenges of designing efficient and specific CRISPR gRNAsIntegrated DNA Technologies
Use of CRISPR-Cas9 has revolutionized targeted genome editing. However, rapid design of high-quality guide RNA (gRNA) sequences with high on-target and low off-target editing remains challenging. We implemented a machine learning algorithm to design high-quality gRNA sequences in 5 commonly used species (human, mouse, rat, zebrafish, and nematode). Our tool also designs gRNA sequences against custom targets, and can check existing gRNA designs for quality. In this webinar, we review our data illustrating this tool's performance and demonstrate its use in predicting and designing improved gRNAs for genome editing. Overview of Single-Cell RNA-seq

Today it is possible to obtain genome-wide transcriptome data from single cells using high-throughput sequencing (scRNA-seq). The main advantage of scRNA-seq is that the cellular resolution and the genome wide scope makes it possible to address issues that are intractable using other methods, e.g. bulk RNA-seq or single-cell RT-qPCR. However, to analyze scRNA-seq data, novel methods are required and some of the underlying assumptions for the methods developed for bulk RNA-seq experiments are no longer valid.
next generation sequemcing

ngs or next generation is and advnced yechnique for determining the sequence of nucleotides in the dna.
Real Time PCR

Real Time Polymerase Chain Reaction
Basics of Real Time PCR
Definition
Advantages
Principles
Instruments (Thermal Cyclers)
Useful terms
Real Time PCR Chemistry
Fluorescence Dyes
SYBR Green
EvaGreen
Melt Doctor
Fluorescence Probes
TaqMan Probe
Molecular Beacons
Scorpion Primers
SYBR Green In details
qPCR Set-Up
Assay Design
Data Analysis
Troubleshooting
A short introduction to single-cell RNA-seq analyses

Groupe de travail Biopuces http://www.nathalievialaneix.eu/biopuces
January 17, 2019
INRA, Toulouse
Digital Droplet PCR

It will give idea about Digital Droplet PCR. Very Useful PPT. It prepared from so many research papers and books
Next Generation Sequencing - the basics

This presentation covers the basics on what Next Generational Sequencing is and some terminology needed in order to understand the process.
Real-Time PCR

Real-time PCR is the continuous collection of fluorescent signal
from one or more polymerase chain reactions over a range of cycles.
The New Xpert MTB/RIF Ultra:

Improving Detection of Mycobacterium Tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing
Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...

Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...Thermo Fisher Scientific
Study of genetic Information from cell-free (cf) DNA provide valuable opportunities in cancer research and potentially impact future oncology. As an example, liquid biopsy provide a non-invasive and cost effective solution for future compared to traditional biopsy tests. Here we report the application of research based Ion Torrent™ next-generation sequencing (NGS) Oncomine™ cfDNA assays and associated workflow, which is developed to detect somatic variants at low frequency of 0.1% in cfDNA from plasma.An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...

An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...Thermo Fisher Scientific
Recent studies in non-invasive biomarker research have demonstrated the potential of using cell-free nucleic acids isolated from blood plasma to serve as surrogates for solid tumors. Somatic mutations representing the tumors could be successfully detected from cell-free DNA (cfDNA) and cell-free RNA (cfRNA), providing new tumor assessment methods in addition to tissue biopsy. However, the low amount of circulating tumor fragments in the blood presents significant challenges for accurate variant detection with NGS assays. Moreover, utilization of both cfDNA and cfRNA requires methods capable of interrogating both types of analytes to maximize the utility of each blood sample.FFPE Applications Solutions brochure

A Platform of comprehensive genomics solutions for analyzing FFPE archival tissues from whole-genome to single molecules
More Related Content
What's hot
Exome seuencing (steps, method, and applications)

Exome sequencing, steps, Next generation sequencing
Rna seq pipeline

RNA Sequence data analysis,Transcriptome sequencing, Sequencing steady state RNA in a sample is known as RNA-Seq. It is free of limitations such as prior knowledge about the organism is not required.
RNA-Seq is useful to unravel inaccessible complexities of transcriptomics such as finding novel transcripts and isoforms.
Data set produced is large and complex; interpretation is not straight forward.
Pcr troubleshooting presentation

Presentation is about the pcr troubles which lead to false results..
Molecular Diagnostics.pptx

Molecular diagnostics is widely used for diagnosing genetic diseases and also the pathogenic diseases by using molecular tools.
Overcoming the challenges of designing efficient and specific CRISPR gRNAs

Overcoming the challenges of designing efficient and specific CRISPR gRNAsIntegrated DNA Technologies
Use of CRISPR-Cas9 has revolutionized targeted genome editing. However, rapid design of high-quality guide RNA (gRNA) sequences with high on-target and low off-target editing remains challenging. We implemented a machine learning algorithm to design high-quality gRNA sequences in 5 commonly used species (human, mouse, rat, zebrafish, and nematode). Our tool also designs gRNA sequences against custom targets, and can check existing gRNA designs for quality. In this webinar, we review our data illustrating this tool's performance and demonstrate its use in predicting and designing improved gRNAs for genome editing. Overview of Single-Cell RNA-seq

Today it is possible to obtain genome-wide transcriptome data from single cells using high-throughput sequencing (scRNA-seq). The main advantage of scRNA-seq is that the cellular resolution and the genome wide scope makes it possible to address issues that are intractable using other methods, e.g. bulk RNA-seq or single-cell RT-qPCR. However, to analyze scRNA-seq data, novel methods are required and some of the underlying assumptions for the methods developed for bulk RNA-seq experiments are no longer valid.
next generation sequemcing

ngs or next generation is and advnced yechnique for determining the sequence of nucleotides in the dna.
Real Time PCR

Real Time Polymerase Chain Reaction
Basics of Real Time PCR
Definition
Advantages
Principles
Instruments (Thermal Cyclers)
Useful terms
Real Time PCR Chemistry
Fluorescence Dyes
SYBR Green
EvaGreen
Melt Doctor
Fluorescence Probes
TaqMan Probe
Molecular Beacons
Scorpion Primers
SYBR Green In details
qPCR Set-Up
Assay Design
Data Analysis
Troubleshooting
A short introduction to single-cell RNA-seq analyses

Groupe de travail Biopuces http://www.nathalievialaneix.eu/biopuces
January 17, 2019
INRA, Toulouse
Digital Droplet PCR

It will give idea about Digital Droplet PCR. Very Useful PPT. It prepared from so many research papers and books
Next Generation Sequencing - the basics

This presentation covers the basics on what Next Generational Sequencing is and some terminology needed in order to understand the process.
Real-Time PCR

Real-time PCR is the continuous collection of fluorescent signal
from one or more polymerase chain reactions over a range of cycles.
The New Xpert MTB/RIF Ultra:

Improving Detection of Mycobacterium Tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing
Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...

Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...Thermo Fisher Scientific
Study of genetic Information from cell-free (cf) DNA provide valuable opportunities in cancer research and potentially impact future oncology. As an example, liquid biopsy provide a non-invasive and cost effective solution for future compared to traditional biopsy tests. Here we report the application of research based Ion Torrent™ next-generation sequencing (NGS) Oncomine™ cfDNA assays and associated workflow, which is developed to detect somatic variants at low frequency of 0.1% in cfDNA from plasma.What's hot (20)
Overcoming the challenges of designing efficient and specific CRISPR gRNAs

Overcoming the challenges of designing efficient and specific CRISPR gRNAs
A short introduction to single-cell RNA-seq analyses

A short introduction to single-cell RNA-seq analyses
Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...

Ion Torrent™ Next Generation Sequencing-Oncomine™ Lung cfDNA assay detected 0...
Similar to Analytical Validation of the Oncomine™ Comprehensive Assay v3 with FFPE and Cell Line Tumor Specimens in a CAP-Accredited and CLIA-Certified Clinical Laboratory
An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...

An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...Thermo Fisher Scientific
Recent studies in non-invasive biomarker research have demonstrated the potential of using cell-free nucleic acids isolated from blood plasma to serve as surrogates for solid tumors. Somatic mutations representing the tumors could be successfully detected from cell-free DNA (cfDNA) and cell-free RNA (cfRNA), providing new tumor assessment methods in addition to tissue biopsy. However, the low amount of circulating tumor fragments in the blood presents significant challenges for accurate variant detection with NGS assays. Moreover, utilization of both cfDNA and cfRNA requires methods capable of interrogating both types of analytes to maximize the utility of each blood sample.FFPE Applications Solutions brochure

A Platform of comprehensive genomics solutions for analyzing FFPE archival tissues from whole-genome to single molecules
Hotspot mutation and fusion transcript detection from the same non-small cell...

Hotspot mutation and fusion transcript detection from the same non-small cell...Thermo Fisher Scientific
The presence of certain chromosomal Header
rearrangements and the subsequent fusion
gene derived from translocations has been
implicated in a number of cancers. Hundreds of
translocations have been described in the
literature recently but the need to efficiently
detect and further characterize these
chromosomal translocations is growing
exponentially. The two main methods to identify
and monitor translocations, fluorescent in situ
hybridization (FISH) and comparative genomic
hybridization (CGH) are challenging, labor
intensive, the information obtained is limited,
and sensitivity is rather low. Common sample
types for these analyses are biopsies or small
tumors, which are very limited in material
making the downstream measurement of more
than one analyte rather difficult; obtaining
another biopsy, using a different section or
splitting the sample can raise issues of tumor
heterogeneity. The ability to study mutation
status as well as measuring fusion transcript
expression from the same sample is powerful
because you’re maximizing the information
obtained from a single precious sample and
eliminating any sample to sample variation.
Here we describe the efficient isolation of two
valuable analytes, RNA and DNA, from the
same starting sample without splitting, followed
by versatile and informative downstream
analysis. This methodology has been applied to
FFPE and degraded samples as well as fresh
tissues, cells and blood. DNA and RNA were
recovered from the same non-small cell lung
adenocarcinoma sample and both mutation
analysis, as well as fusion transcript detection
was performed using the Ion Torrent PGM™
platform on the same Ion 318™ chip. Using
10ng of DNA and 10ng of RNA input, we
applied the Ion AmpliSeq™ Colon and Lung
Cancer panel to analyze over 500 COSMIC
mutations in 22 genes and the Ion AmpliSeq™
RNA Lung Fusion panel to detect 40 different
fusion transcripts.Comparing Mutation Detection Sensitivity from Matched FFPE Tissue and Liquid ...

Comparing Mutation Detection Sensitivity from Matched FFPE Tissue and Liquid ...Thermo Fisher Scientific
Cancer researchers are avidly working to enable circulating cell free DNA (cfDNA) profiling as a new more sensitive tool to detect and screen for the presence of solid tumors before detection through clinical methods. Despite the high level of interest in cfDNA, researchers still have reservations until enough data has demonstrated complementarity between methodologies. In this study, we examined the data quality and concordance of mutations called for a small number of matched formalin fixed paraffin embedded (FFPE) tissue and plasma samples.Analytical performance of a novel next generation sequencing assay for Myeloi...

Analytical performance of a novel next generation sequencing assay for Myeloi...Thermo Fisher Scientific
To support clinical and translational research into precision oncology strategies for myeloid cancers, a next-generation sequencing (NGS) assay was developed to detect common and relevant somatic alterations. To define gene targets that were recurrently altered in myeloid cancers and relevant for clinical and translational research, an extensive survey of investigators at hematology oncology research labs was performed.Pharmacogenomics Research Ion AmpliSeq Assay | ESHG 2015 Poster PM15.10

Cytochrome P450 enzymes metabolize about 75% of drugs, including oncology drugs, with UGT enzymes metabolizing about another 15%. Variations in gene sequence or in copy number may result in an inactive, defective, unstable, mis-spliced, low expressed, or absent enzyme, an increase in enzyme activity, or an altered affinity for substrates. Pharmacogenomics genes can predict whether an individual is a poor or rapid metabolizer, facilitating dose optimization. Failure to adjust dosage of drugs metabolized by the relevant enzyme can lead to adverse drug reaction, or conversely to too rapid drug metabolism and no drug response. Here, we present a pharmacogenomics (PGx) Research panel to detect 139 SNV/Indel targets in 42 genes (Figure 1) and CYP2D6 copy number variation (CNV, Figure 2). This panel covers the commonly known targets in genes encoding drug metabolism enzymes and associated transport proteins. The panel design is particularly challenging due to high levels of sequence homology between the cytochrome P450 genes. This assay uses Ion AmpliSeq™ technology and contains 146 amplicons in an ultrahigh multiplex PCR in a single pool, followed by Ion Torrent™ semiconductor sequencing. The assay requires as little as 10 ng of input DNA. This customizable panel allows target addition or removal, and will be the first NGS PGx Research panel on the market.
Tools for lncRNA research in cancer

The human genome is pervasively transcribed, giving rise to an increasing number of long non-coding RNA genes. Most of these genes are novel or poorly characterized, and their relevance in human health and disease remains elusive. In our lab, we have developed various tools to study lncRNAs, amongst others to assess their role in cancer. As such, we are looking for novel biomarkers and therapeutic targets. I will describe various tools and ongoing research programs, including a comprehensive annotated catalog of human lncRNAs (LNCipedia), a targeted screen for focal lncRNA copy number alterations, a web tool for antisense oligonucleotide design, Zipper plot to visualize the transcriptional activity of lncRNAs in their genomic context, decodeRNA functional context mapping, and probe based lncRNA capture sequencing in body fluids.
Tumor Mutational Load assessment of FFPE samples using an NGS based assay

Understanding the molecular determinants of response to immune checkpoint blockade inhibitors is a critical unmet need for translational oncology research. Research tools to characterize the mutational landscape of cancers may potentially help identify predictive biomarkers for immuno-therapy that can be tested in future studies. Herein, we describe a targeted Ion AmpliSeq assay to determine the mutational load and signature of cancer research samples.
Development of Quality Control Materials for Characterization of Comprehensiv...

Development of Quality Control Materials for Characterization of Comprehensiv...Thermo Fisher Scientific
Targeted next-generation sequencing (NGS) panels can detect hundreds of mutations in key genes using amplification based and hybrid-capture based NGS technologies. Although NGS technology is a powerful tool, optimizing and characterizing test performance on hundreds of variants is extremely challenging, time consuming, and expensive. Samples must be sourced, variants identified and orthogonally confirmed, then quantified and diluted. This effort is then multiplied across dozens of samples, and then samples must be run over many runs and days to assess assay reproducibility, precision, sensitivity, etc. In this study, we developed a novel reference material, experimental design, and analysis pipeline that allows for highly streamlined NGS assay characterization, enabling thorough test characterization across 500+ variants within only 6 runs.Ion Torrent™ Next Generation Sequencing – Detect 0.1% Low Frequency Somatic V...

Ion Torrent™ Next Generation Sequencing – Detect 0.1% Low Frequency Somatic V...Thermo Fisher Scientific
Accurate detection of low-frequency somatic mutations as well as low level structural variants such as copy number variation (CNV) in circulating cell-free DNA (cfDNA) using blood samples from subjects previously diagnosed with cancer provides a potential non-invasive approach to monitor cancer status and evaluate cancer evolution in the future. We have previously reported the Oncomine™ Breast cfDNA Assay enables detection of somatic mutations in plasma down to a level of 0.1% variant allelic frequency in breast cancer relevant genes. Here we extend this technology to simultaneously detect single nucleotide variants (SNVs) as well as copy number variation (CNV) from a single cfDNA sample.Similar to Analytical Validation of the Oncomine™ Comprehensive Assay v3 with FFPE and Cell Line Tumor Specimens in a CAP-Accredited and CLIA-Certified Clinical Laboratory (20)
An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...

An Efficient NGS Workflow for Liquid Biopsy Research Using a Comprehensive As...
Hotspot mutation and fusion transcript detection from the same non-small cell...

Hotspot mutation and fusion transcript detection from the same non-small cell...
Comparing Mutation Detection Sensitivity from Matched FFPE Tissue and Liquid ...

Comparing Mutation Detection Sensitivity from Matched FFPE Tissue and Liquid ...
2014-Yeo-A Multiplex Two-Color Real-Time PCR Method(1)

2014-Yeo-A Multiplex Two-Color Real-Time PCR Method(1)
Analytical performance of a novel next generation sequencing assay for Myeloi...

Analytical performance of a novel next generation sequencing assay for Myeloi...
Pharmacogenomics Research Ion AmpliSeq Assay | ESHG 2015 Poster PM15.10

Pharmacogenomics Research Ion AmpliSeq Assay | ESHG 2015 Poster PM15.10
Services related to Genetics _ Genetic Testing.pptx

Services related to Genetics _ Genetic Testing.pptx
Personalized Oncology Through Integrative High-Throughput Sequencing:

Personalized Oncology Through Integrative High-Throughput Sequencing:
Tumor Mutational Load assessment of FFPE samples using an NGS based assay

Tumor Mutational Load assessment of FFPE samples using an NGS based assay
Development of Quality Control Materials for Characterization of Comprehensiv...

Development of Quality Control Materials for Characterization of Comprehensiv...
Ion Torrent™ Next Generation Sequencing – Detect 0.1% Low Frequency Somatic V...

Ion Torrent™ Next Generation Sequencing – Detect 0.1% Low Frequency Somatic V...
NUGEN-X-Gen_2011_poster_trancriptome_sequencing_RNA-Seq

NUGEN-X-Gen_2011_poster_trancriptome_sequencing_RNA-Seq
More from Thermo Fisher Scientific
Why you would want a powerful hot-start DNA polymerase for your PCR

Hot-start DNA polymerases are commonly used in PCR for genotyping, sequencing, molecular diagnostics, and high-throughput applications. In this presentation, PCR performance of Invitrogen™ Platinum II Taq Hot-Start DNA Polymerase and Invitrogen™ AccuPrime Taq DNA Polymerase is compared in the following areas:
• PCR run time for targets of different lengths
• Amplification of AT-rich and GC-rich sequences
• Tolerance to PCR inhibitors
• Sensitivity in target detection
• Universal protocol for PCR targets of different lengths
• Multiplex PCR of 15 targets
• Product format for direct gel loading
Request a sample of Platinum II Taq enzyme at http://bit.ly/2M4U9cw
Find other PCR enzymes at http://bit.ly/2JIPrzj
Learn more about PCR at http://bit.ly/2y2aSVo
#PCR #PCREducation #Invitrogen #InvitrogenSchoolofMolBio
TCRB chain convergence in chronic cytomegalovirus infection and cancer

Human cytomegalovirus (CMV) is a common immune-evasive herpes family virus leading to lifelong asymptomatic infection in 50 to 80% of humans. Current research evaluating the use of
TCR sequencing to predict response to immunotherapy has focused on measurements of T cell clonal expansion and TCR convergence (2,3,4) as potential predictive biomarkers for
response. Given that CMV infection has been reported to elicit large clonal proliferations of CMV reactive T cells (1), and is a source of chronic antigen stimulation, we hypothesized that CMV
infection might alter T cell repertoire features in a manner relevant to the potential biomarker use of TCR sequencing. Here we sought to identify features of CMV infection using TCRB profiling of
peripheral blood (PBL) total RNA. We identify reduced T cell evenness and elevated TCR convergence as features of chronic CMV infection.
Improvement of TMB Measurement by removal of Deaminated Bases in FFPE DNA

Tumor mutational burden (TMB) is a positive predictive factor for response to immune-checkpoint inhibitors in certain types of cancer. The Oncomine™ Tumor Mutation Load Assay, a targeted next generation sequencing (NGS) assay, measures TMB (from 1.2Mb of coding region) and detects mutations in 409 cancer genes. The TMB values obtained using targeted sequencing are highly correlated with TMB measured by whole exome sequencing. FFPE preservation methods can lead to significant cytosine deamination of the isolated DNA, resulting in decreased sequencing quality. In these samples, uracils are propagated as thymines and result in false C>T substitutions. Analysis of the Oncomine™ TML Assay using Torrent Suite and Ion Reporter ™ software uniquely estimates the degree of deamination in fixed tissues by measuring C:G>T:A variants. This deamination score is used to assess quality of DNA extracted from FFPE tumor tissue. To minimize the influence
that excess deamination has on TMB results, we have incorporated a repair treatment to eliminate damaged targets and improve usable TMB values of DNA from damaged FFPE tumor tissue using Uracil-DNA glycosylase (UDG). The
Oncomine™ TML Assay for TMB on the Ion Gene Studio™ S5 systems in conjunction with a deamination score is informative and potentially predictive for the use of checkpoint inhibitors in multiple cancer types.
What can we learn from oncologists? A survey of molecular testing patterns

Oncologists are increasingly incorporating NGS testing to guide targeted and immuno-oncology therapies1. Most clinical NGS testing is confined to large academic institutions and reference labs, despite the fact that most cancer patients are treated in the community settings. We therefore sought to examine molecular testing selection patterns directly from oncologists in order to better identify perceived gaps in testing and treatment paradigms
Evaluation of ctDNA extraction methods and amplifiable copy number yield usin...

Evaluation of ctDNA extraction methods and amplifiable copy number yield usin...Thermo Fisher Scientific
The use of cell-free circulating tumor DNA (ctDNA) for non-invasive cancer testing has the potential to revolutionize the field. However, emergence of an increasing number of extraction methods and detection assays is rendering laboratory workflow development much more complex and cumbersome. The use of standardized, well characterized ctDNA control materials in human plasma could facilitate the evaluation of extraction efficiency and assay performance across platforms. In this study, we use a full process ctDNA quality control material in true human plasma to demonstrate the variability of extraction yield between different ctDNA extraction kits. We also examine the correlation between the amplifiable
copy number and DNA concentration post-extraction.Novel Spatial Multiplex Screening of Uropathogens Associated with Urinary Tra...

Novel Spatial Multiplex Screening of Uropathogens Associated with Urinary Tra...Thermo Fisher Scientific
Accurate identification of uropathogens in a timely manner is important to correctly understand urinary tract infections(UTI’s), which affects nearly 150 million people each year. The
current standard approach for detecting the UTI pathogens is culture based. This method is time consuming, has low throughput, and can lack sensitivity and/or specificity. In addition, not all uropathogens grow equally well under standard culture conditions which can result in a failure to detect the species. To address these gaps, we have developed a unique workflow from sample preparation to target identification using the nanofluidic OpenArray™ platform for spatial multiplexing of target specific assays. In this study, we tested pre-determined blinded research samples and confirmed the subset of results with orthogonal Sanger sequences.Liquid biopsy quality control – the importance of plasma quality, sample prep...

Liquid biopsy quality control – the importance of plasma quality, sample prep...Thermo Fisher Scientific
Liquid biopsy is emerging as a non-invasive companion to traditional solid tumor biopsies. As next generation sequencing (NGS) of circulating cell-free nucleic acids (cfNA = cfDNA and cfRNA) becomes common, it’s important to understand the impact of sample preparation on quality, specificity, and sensitivity of liquid biopsy tests. Plasma samples are often limited, and may have undesirable characteristics such as lipemia or hemolysis that contribute unwanted genomic DNA (gDNA) to the sample. Low cfDNA concentration can also limit the amount available for NGS library prep. In this study, we explore the effects of suboptimal plasma and low library input on liquid biopsy NGS, and discuss various techniques for in-process quality control of cfNA samples isolated from plasmaStreamlined next generation sequencing assay development using a highly multi...

Streamlined next generation sequencing assay development using a highly multi...Thermo Fisher Scientific
Next generation sequencing (NGS) assay development for solid tumor sequencing requires characterization of variant calling directly from formalin-fixed paraffin embedded (FFPE) tissue samples. However, cell line based FFPE and human FFPE samples only contain 2 to 20 variants, which require laboratories to invest significant resources in sample sourcing and preparation when developing assays to detect 100+ variantsTargeted T-cell receptor beta immune repertoire sequencing in several FFPE ti...

Targeted T-cell receptor beta immune repertoire sequencing in several FFPE ti...Thermo Fisher Scientific
T-cell receptor beta (TCRβ) immune repertoire analysis by next-generation sequencing is a valuable tool for studies of the tumor microenvironment and potential immune responses to cancer immunotherapy. Here we describe a TCRβ sequencing assay that leverages the low sample input requirements of AmpliSeq library preparation technology to extend the capability of targeted immune repertoire sequencing to include FFPE samples which can often be degraded and in short supplyA High Throughput System for Profiling Respiratory Tract Microbiota

As one of the leading causes of death globally, respiratory
infections could be caused by single or multiple types of viral,
bacterial or fungal pathogens that present in the upper and
lower respiratory tract. Panel-based testing using molecular
methods to identify multiple pathogens simultaneously can
contribute to better understanding of respiratory infections.
A high-throughput approach for multi-omic testing for prostate cancer research

A high-throughput approach for multi-omic testing for prostate cancer researchThermo Fisher Scientific
The proliferation of genetic testing technologies and genome-scale studies has increased our understanding of the genetic basis of complex diseases. However, this information alone tells an incomplete story of the underlying biology. Integrative approaches that combine data from multiple sources, such as the genome, transcriptome and/or proteome, can provide a more comprehensive and multi-dimensional model of complex diseases. Similarly, the integration of multiple data types in disease screening can improve our understanding of disease in populations. In a series of groundbreaking multi-omic, population-based studies of prostate cancer, researchers at the Karolinska Institutet in Stockholm, Sweden identified sets of genetic and protein biomarkers that when evaluated together with other clinical research data performed significantly better in predicting cancer risk (1,2) than the most-widely used single protein biomarker, the prostate-specific antigen (PSA).Why is selecting the right thermal cycler important?

Discover the innovations and more that led to amazing discoveries through the use of thermal cyclers. What were scientists able to accomplish? What things are important to them when selecting a thermal cycler? What do you need to advance your science?
Learn more about thermal cyclers: http://bit.ly/2Q2oPhF
See all thermal cycler offerings: http://bit.ly/2Paf1wH
A rapid library preparation method with custom assay designs for detection of...

A rapid library preparation method with custom assay designs for detection of...Thermo Fisher Scientific
Herein, we describe a new research method for library
preparation using the Ion AmpliSeq™ HD Library Kit with
custom assay designs from Ion AmpliSeq HD Panels for
detection of low level variants from liquid biopsy samples. This
method includes incorporation of molecular tags that enable
0.1% Limit of Detection (LOD) in cell free DNA (cfDNA) and
dual barcodes for sample identification. This method is also
applicable to formalin-fixed paraffin embedded (FFPE)
samples. The libraries can be prepared in as little as 3 hours
and are compatible for analysis with the Ion GeneStudio™ S5
systemGeneration of Clonal CRISPR/Cas9-edited Human iPSC Derived Cellular Models an...

Generation of Clonal CRISPR/Cas9-edited Human iPSC Derived Cellular Models an...Thermo Fisher Scientific
Reprogramming permits the derivation of hiPSCs from diseased patients, and allows us to model diseases in vitro. Furthermore, with the advent of CRISPR mediated genome editing, we can now mimic disease mutations in control hiPSC lines to study the biological effect of just those mutations. hiPSCs can then be differentiated into specified cell types such as neurons which can be used to develop assays for drug safety screening or can be used to model disease phenotypes in a dish to discover new drugs.TaqMan®Advanced miRNA cDNA synthesis kit to simultaneously study expression o...

TaqMan®Advanced miRNA cDNA synthesis kit to simultaneously study expression o...Thermo Fisher Scientific
MicroRNAs (miRNA) are a class of small non-coding RNAs (approximately 21 nt long) that bind complementary sequences in target mRNAs to specifically regulate gene expression. Aberrant regulation of miRNAs and their targets has been associated with several diseases including cancer. The relationship between miRNA and mRNA has been found to be important in cancer development and progression. Simultaneous expression studies of miRNA and mRNA and detection of mutations in mRNA transcripts can be valuable in understanding molecular mechanisms that
have an underlying role in various diseases. We demonstrate the technical verification of a novel method to reverse-transcribe and pre-amplify miRNA and mRNA from sample-limiting serum research samples using the TaqMan® Advanced miRNA cDNA Synthesis Kit. Based on results from previous studies, a signature of 49 mRNA and 37 miRNA targets has been identified that may help distinguish between benign and malignant pancreatic tissues. In this study, these targets and an additional set of transcript mutations were analyzed in serum from normal and test samples. TaqMan assays for miRNA and mRNA targets and custom TaqMan Mutation Detection Assays (TMDAs) were placed on TaqMan Array Cards to facilitate investigation of several samples in a single experiment. Results demonstrate that transcript mutations can be detected and miRNA and mRNA targets can be reliably quantified from a single reverse transcription reaction. For research use only. Not for use in diagnostic purposes.Identifying novel and druggable targets in a triple negative breast cancer ce...

Identifying novel and druggable targets in a triple negative breast cancer ce...Thermo Fisher Scientific
In this study, we developed a CRISPR/Cas9-based high throughput loss-of-function screen for identifying target genes responsible for the tumor proliferation and growth in TNBC. Our initial focus was to identify essential kinases in MDA-MB-231 cell line using the Invitrogen™ LentiArray™ Human Kinase CRISPR Library, which targets 840 kinases with up to 4 different gRNAs per protein kinase for complete gene knockout. This functional screen identified over 90 protein kinases that are essential for cell viability and cell proliferation. Ten of these hits (CDK1, CDK2, CDK8, CDK10, CDK11A, CDK19, CDK19, CDC7, EPHA2 and WEE1) are well-known targets validated in the literature. Currently, we are in the process validating the novel hits through target gene sequencing, western blotting and target specific small molecule kinase inhibitors.Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrati...

Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrati...Thermo Fisher Scientific
T cell convergence refers to the phenomenon whereby antigen-driven selection enriches for T cell receptors (TCRs) having a shared antigen specificity but different amino acid or
nucleotide sequence. T cell recruitment and expansion within the tumor microenvironment (TME) may be directed by responses to tumor neoantigen, suggesting that elevated T
cell convergence could be a general feature of the tumor infiltrating T cell repertoire. Here we use the Ion AmpliSeq™ Immune Repertoire Assay Plus – TCRβ to evaluate evidence
for T cell convergence within melanoma tumor biopsy research samples from a set of 63 subjects plus peripheral blood leukocytes (PBL) from four healthy subjects. We find that the melanoma TME is highly enriched for convergent TCRs compared to healthy donor peripheral blood. We discuss the potential use of TCR convergence as a liquid biopsy compatible predictive biomarker for immunotherapy response.Estimating Mutation Load from Tumor Research Samples using a Targeted Next-Ge...

Estimating Mutation Load from Tumor Research Samples using a Targeted Next-Ge...Thermo Fisher Scientific
Tumor mutation load predicts durable benefit from immune checkpoint inhibitors in several cancer types. Existing methods to estimate tumor mutation load have large input DNA and extensive infrastructure requirements and are associated with delays due to shipping biopsy samples to central laboratories. We demonstrate the ability of a targeted panel with fast
turn-around time and low input requirement for estimating mutation load from tumor samples to advance research in immuno-oncology.Development of a next-generation (NGS) assay for pediatric, childhood, and yo...

Development of a next-generation (NGS) assay for pediatric, childhood, and yo...Thermo Fisher Scientific
The study of recurrent somatic alterations associated with pediatric, childhood and young adult cancers has lagged behind those that associated with adult cancers. Whole exome and transcriptome approaches are still being used to support discovery efforts, consequently, due to several initiatives aimed at profiling genomic alterations associated with childhood cancers, a set of recurrent somatic alterations has been defined.High content screening in MCF7 and MDA-MB231 cells show differential response...

High content screening in MCF7 and MDA-MB231 cells show differential response...Thermo Fisher Scientific
Oxygen levels in typical cell culture conditions do not accurately reflect the oxygen levels cells are exposed to within the body. Furthermore, oxygen levels can vary within the tumor microenvironment. These variances can affect how cells respond to a variety of drugs and small molecules. To further understand how oxygen levels affect drug sensitivity, the response of hormone-dependent MCF7 cells were compared to hormone-independent MDA-MB231 cells, cultured under low and high oxygen.More from Thermo Fisher Scientific (20)
Why you would want a powerful hot-start DNA polymerase for your PCR

Why you would want a powerful hot-start DNA polymerase for your PCR
TCRB chain convergence in chronic cytomegalovirus infection and cancer

TCRB chain convergence in chronic cytomegalovirus infection and cancer
Improvement of TMB Measurement by removal of Deaminated Bases in FFPE DNA

Improvement of TMB Measurement by removal of Deaminated Bases in FFPE DNA
What can we learn from oncologists? A survey of molecular testing patterns

What can we learn from oncologists? A survey of molecular testing patterns
Evaluation of ctDNA extraction methods and amplifiable copy number yield usin...

Evaluation of ctDNA extraction methods and amplifiable copy number yield usin...
Novel Spatial Multiplex Screening of Uropathogens Associated with Urinary Tra...

Novel Spatial Multiplex Screening of Uropathogens Associated with Urinary Tra...
Liquid biopsy quality control – the importance of plasma quality, sample prep...

Liquid biopsy quality control – the importance of plasma quality, sample prep...
Streamlined next generation sequencing assay development using a highly multi...

Streamlined next generation sequencing assay development using a highly multi...
Targeted T-cell receptor beta immune repertoire sequencing in several FFPE ti...

Targeted T-cell receptor beta immune repertoire sequencing in several FFPE ti...
A High Throughput System for Profiling Respiratory Tract Microbiota

A High Throughput System for Profiling Respiratory Tract Microbiota
A high-throughput approach for multi-omic testing for prostate cancer research

A high-throughput approach for multi-omic testing for prostate cancer research
Why is selecting the right thermal cycler important?

Why is selecting the right thermal cycler important?
A rapid library preparation method with custom assay designs for detection of...

A rapid library preparation method with custom assay designs for detection of...
Generation of Clonal CRISPR/Cas9-edited Human iPSC Derived Cellular Models an...

Generation of Clonal CRISPR/Cas9-edited Human iPSC Derived Cellular Models an...
TaqMan®Advanced miRNA cDNA synthesis kit to simultaneously study expression o...

TaqMan®Advanced miRNA cDNA synthesis kit to simultaneously study expression o...
Identifying novel and druggable targets in a triple negative breast cancer ce...

Identifying novel and druggable targets in a triple negative breast cancer ce...
Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrati...

Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrati...
Estimating Mutation Load from Tumor Research Samples using a Targeted Next-Ge...

Estimating Mutation Load from Tumor Research Samples using a Targeted Next-Ge...
Development of a next-generation (NGS) assay for pediatric, childhood, and yo...

Development of a next-generation (NGS) assay for pediatric, childhood, and yo...
High content screening in MCF7 and MDA-MB231 cells show differential response...

High content screening in MCF7 and MDA-MB231 cells show differential response...
Recently uploaded
insect taxonomy importance systematics and classification

documents provide information about insect classification and taxonomy of insect
Predicting property prices with machine learning algorithms.pdf

This is a report about predicting property prices using SVM, random forest and gradient boosting machine
Citrus Greening Disease and its Management

Citrus Greening was one of the major causes of decline in the citrus production. So, effective management cultural practices should be incorporated
Seminar of U.V. Spectroscopy by SAMIR PANDA

Spectroscopy is a branch of science dealing the study of interaction of electromagnetic radiation with matter.
Ultraviolet-visible spectroscopy refers to absorption spectroscopy or reflect spectroscopy in the UV-VIS spectral region.
Ultraviolet-visible spectroscopy is an analytical method that can measure the amount of light received by the analyte.
Richard's aventures in two entangled wonderlands

Since the loophole-free Bell experiments of 2020 and the Nobel prizes in physics of 2022, critics of Bell's work have retreated to the fortress of super-determinism. Now, super-determinism is a derogatory word - it just means "determinism". Palmer, Hance and Hossenfelder argue that quantum mechanics and determinism are not incompatible, using a sophisticated mathematical construction based on a subtle thinning of allowed states and measurements in quantum mechanics, such that what is left appears to make Bell's argument fail, without altering the empirical predictions of quantum mechanics. I think however that it is a smoke screen, and the slogan "lost in math" comes to my mind. I will discuss some other recent disproofs of Bell's theorem using the language of causality based on causal graphs. Causal thinking is also central to law and justice. I will mention surprising connections to my work on serial killer nurse cases, in particular the Dutch case of Lucia de Berk and the current UK case of Lucy Letby.
extra-chromosomal-inheritance[1].pptx.pdfpdf![extra-chromosomal-inheritance[1].pptx.pdfpdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![extra-chromosomal-inheritance[1].pptx.pdfpdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Slide 1: Title Slide
Extrachromosomal Inheritance
Slide 2: Introduction to Extrachromosomal Inheritance
Definition: Extrachromosomal inheritance refers to the transmission of genetic material that is not found within the nucleus.
Key Components: Involves genes located in mitochondria, chloroplasts, and plasmids.
Slide 3: Mitochondrial Inheritance
Mitochondria: Organelles responsible for energy production.
Mitochondrial DNA (mtDNA): Circular DNA molecule found in mitochondria.
Inheritance Pattern: Maternally inherited, meaning it is passed from mothers to all their offspring.
Diseases: Examples include Leber’s hereditary optic neuropathy (LHON) and mitochondrial myopathy.
Slide 4: Chloroplast Inheritance
Chloroplasts: Organelles responsible for photosynthesis in plants.
Chloroplast DNA (cpDNA): Circular DNA molecule found in chloroplasts.
Inheritance Pattern: Often maternally inherited in most plants, but can vary in some species.
Examples: Variegation in plants, where leaf color patterns are determined by chloroplast DNA.
Slide 5: Plasmid Inheritance
Plasmids: Small, circular DNA molecules found in bacteria and some eukaryotes.
Features: Can carry antibiotic resistance genes and can be transferred between cells through processes like conjugation.
Significance: Important in biotechnology for gene cloning and genetic engineering.
Slide 6: Mechanisms of Extrachromosomal Inheritance
Non-Mendelian Patterns: Do not follow Mendel’s laws of inheritance.
Cytoplasmic Segregation: During cell division, organelles like mitochondria and chloroplasts are randomly distributed to daughter cells.
Heteroplasmy: Presence of more than one type of organellar genome within a cell, leading to variation in expression.
Slide 7: Examples of Extrachromosomal Inheritance
Four O’clock Plant (Mirabilis jalapa): Shows variegated leaves due to different cpDNA in leaf cells.
Petite Mutants in Yeast: Result from mutations in mitochondrial DNA affecting respiration.
Slide 8: Importance of Extrachromosomal Inheritance
Evolution: Provides insight into the evolution of eukaryotic cells.
Medicine: Understanding mitochondrial inheritance helps in diagnosing and treating mitochondrial diseases.
Agriculture: Chloroplast inheritance can be used in plant breeding and genetic modification.
Slide 9: Recent Research and Advances
Gene Editing: Techniques like CRISPR-Cas9 are being used to edit mitochondrial and chloroplast DNA.
Therapies: Development of mitochondrial replacement therapy (MRT) for preventing mitochondrial diseases.
Slide 10: Conclusion
Summary: Extrachromosomal inheritance involves the transmission of genetic material outside the nucleus and plays a crucial role in genetics, medicine, and biotechnology.
Future Directions: Continued research and technological advancements hold promise for new treatments and applications.
Slide 11: Questions and Discussion
Invite Audience: Open the floor for any questions or further discussion on the topic.
Multi-source connectivity as the driver of solar wind variability in the heli...

The ambient solar wind that flls the heliosphere originates from multiple
sources in the solar corona and is highly structured. It is often described
as high-speed, relatively homogeneous, plasma streams from coronal
holes and slow-speed, highly variable, streams whose source regions are
under debate. A key goal of ESA/NASA’s Solar Orbiter mission is to identify
solar wind sources and understand what drives the complexity seen in the
heliosphere. By combining magnetic feld modelling and spectroscopic
techniques with high-resolution observations and measurements, we show
that the solar wind variability detected in situ by Solar Orbiter in March
2022 is driven by spatio-temporal changes in the magnetic connectivity to
multiple sources in the solar atmosphere. The magnetic feld footpoints
connected to the spacecraft moved from the boundaries of a coronal hole
to one active region (12961) and then across to another region (12957). This
is refected in the in situ measurements, which show the transition from fast
to highly Alfvénic then to slow solar wind that is disrupted by the arrival of
a coronal mass ejection. Our results describe solar wind variability at 0.5 au
but are applicable to near-Earth observatories.
SCHIZOPHRENIA Disorder/ Brain Disorder.pdf

This pdf is about the Schizophrenia.
For more details visit on YouTube; @SELF-EXPLANATORY;
https://www.youtube.com/channel/UCAiarMZDNhe1A3Rnpr_WkzA/videos
Thanks...!
Recently uploaded (20)
Circulatory system_ Laplace law. Ohms law.reynaults law,baro-chemo-receptors-...

Circulatory system_ Laplace law. Ohms law.reynaults law,baro-chemo-receptors-...
insect taxonomy importance systematics and classification

insect taxonomy importance systematics and classification
Predicting property prices with machine learning algorithms.pdf

Predicting property prices with machine learning algorithms.pdf
Body fluids_tonicity_dehydration_hypovolemia_hypervolemia.pptx

Body fluids_tonicity_dehydration_hypovolemia_hypervolemia.pptx
Lateral Ventricles.pdf very easy good diagrams comprehensive

Lateral Ventricles.pdf very easy good diagrams comprehensive
PRESENTATION ABOUT PRINCIPLE OF COSMATIC EVALUATION

PRESENTATION ABOUT PRINCIPLE OF COSMATIC EVALUATION
Multi-source connectivity as the driver of solar wind variability in the heli...

Multi-source connectivity as the driver of solar wind variability in the heli...
Analytical Validation of the Oncomine™ Comprehensive Assay v3 with FFPE and Cell Line Tumor Specimens in a CAP-Accredited and CLIA-Certified Clinical Laboratory
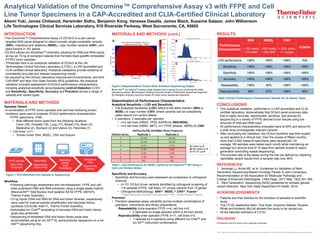
- 1. ©2018 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Determination of Performance Characteristics Analytical Sensitivity – LOD and Sensitivity • The Analytical Sensitivity (LOD) refers to the allelic fraction (SNV or INDEL) or copy number (CNV) where variants can be consistently called above non-variant alleles. • 2 operators, 2 replicates per operator • n=3 cell lines (KRAS, TP53, EGFR) for SNV/INDEL • n=2 cell lines (HER2, MET), n=1 FFPE (Breast, HER2) for CNV Specificity and Accuracy • Specificity and Accuracy were assessed via comparison to orthogonal methods. • n=131 OCAv3 driver variants identified by orthogonal screening of 114 samples (FFPE, cell lines); 27 unique variants from 15 genes • Orthogonal Methodology: SNV▲, INDEL ▲,CNV■, Fusion● ▲ Sanger Sequencing & OncomineTM Focus Assay NGS, ■ FISH, ● qPCR & PervenioTM Lung NGS Precision • Precision assesses assay variability across multiple combinations of operators, instruments and library preparations. Repeatability (intra-operator) FFPE n=4, cell line n=2 • 3 replicates by single operator within a 24 hour period Reproducibility (inter-operator) FFPE n=11, cell lines n=3 • 1 replicate by 4 operators using different Ion ChefTM and Ion S5TM instrument combinations ■ Called by IR ■ Not called by IR 5% AF cutoff FFPE sample DNA+RNA Extraction DNA+RNA Sequencing Library Prep Barcoded Library Chef Templating Templated Chip Sequencing Results INTRODUCTION •The Oncomine™ Comprehensive Assay v3 (OCAv3) is a pan-cancer targeted NGS panel designed to detect somatic single-nucleotide variants (SNV), insertions and deletions (INDEL), copy number variants (CNV), and gene fusions in 161 genes. •OCAv3 utilizes Ion AmpliSeqTM chemistry, allowing for DNA and RNA inputs as low as 10 ng of extracted material from formalin-fixed paraffin embedded (FFPE) tumor samples. • Presented here is an analytical validation of OCAv3 at the Life Technologies Clinical Services Laboratory (LTCSL), a CAP-accredited and CLIA-certified clinical laboratory. Analytical validations provide evidence of consistently accurate and relevant sequencing results. •As required by the Clinical Laboratory Improvement Amendments, and both CAP/AMP and New York State Somatic NGS guidelines, the analytical validation included assessment of OCAv3 performance characteristics, including analytical sensitivity (encompassing Limit-of-Detection (LOD) and Sensitivity), Specificity, Accuracy and Precision across a range of variants and variant types. MATERIALS AND METHODS Samples Tested •A combination of FFPE tumor samples and cell lines harboring known mutations were used to evaluate OCAv3 performance characteristics •FFPE specimens, n=99 • Nine different tumor types from the following locations: Colon (39), Prostate (18), Lung (17), Breast (15), Brain (3), Melanoma (2), Stomach (2) and Uterus (2), Pancreas (1) • Cell lines, n=11 • Known tumor SNV, INDEL, CNV and fusions Workflow •Following pathology assessment and microdissection, FFPE and cell lines underwent DNA and RNA extraction using a single-lysate method (RecoverAllTM Total Nucleic Acid Isolation Kit for FFPE, AM1975, Thermo Fisher Scientific). •10 ng inputs (DNA and RNA for DNA and fusion libraries, respectively) were used for manual sample amplification and barcoded library synthesis (OCAv3M, A36111, Thermo Fisher Scientific) •Automated Ion ChefTM templating of barcoded DNA and fusion library pools was performed. •Sequencing of templated DNA and fusion library pools was accomplished using an Ion S5TM XL semiconductor sequencer on a Ion 540TM sequencing chip. Akemi Yuki, James Chitwood, Harwinder Sidhu, Benjamin Kong, Vanessa Dasalla, James Black, Suzanne Salazar, John Williamson Life Technologies Clinical Services Laboratory, 910 Riverside Parkway, West Sacramento, CA, 95605 Analytical Validation of the Oncomine™ Comprehensive Assay v3 with FFPE and Cell Line Tumor Specimens in a CAP-Accredited and CLIA-Certified Clinical Laboratory Figure 1 OCA Workflow from Sample to Sequencing Figure 2 Representative Torrent Suite Software Sequencing Output A) Ion 540TM Ion SphereTM loading image (loading here is typical of a run producing 80 million sequencing reads). B) Histogram showing count and length of DNA library sequenced fragments. C) Histogram showing count and length of Fusion library sequenced fragments. MATERIALS AND METHODS (cont.) RESULTS CONCLUSIONS • This analytical validation, performed in a CAP-accredited and CLIA- certified laboratory, demonstrates that OCAv3 panel produces data that is highly accurate, reproducible, sensitive, and precise for sequencing in a variety of FFPE-derived tumor tissues using low amounts of DNA and RNA input. • All performance characteristics evaluated were at 90% and above for a wide array of biologically relevant variants • After concluding the validation, the OCAv3 workflow was then scaled up and applied to a clinical trial. Over the course of fifteen months, more than 2,500 research specimens were sequenced. On average,180 samples were tested each month while maintaining an average turn around time of 10 days from sample receipt to report generation (excluding repeat sequencing). • The success rate of the assay during the trial (as defined by obtaining reportable variant results from a sample) was over 95%. REFERENCES 1. Jennings LJ, Arcila ME, et al. Guidelines for Validation of Next- Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017 May; 19(3):341-365 2. “Next Generation” Sequencing (NGS) guidelines for somatic genetic variant detection. New York State Department of Health. 2016. ACKNOWLEDGEMENTS • Patients and their families for the donation of samples to scientific study. • The LTCSL leadership team: Tina Huan, Suzanne Salazar, Roxana White and Kimberly McCall allowed this study to be carried out. • All the talented members of LTCSL. Disclaimer For Research Use Only. Not for use in diagnostic procedures. Performance Characteristic SNV INDEL CNV FUSION> 250 reads > 5% MAF >250 reads > 5% MAF > 50% tumor > 7 copies LOD performance >99% >99% >99% N/A Sensitivity >99% >99% >99% >99% Specificity >99% >99% >99% >99% Accuracy >99% >99% >99% >99% Repeatability >99% 90% >99% >99% Reproducibility 96% 97% >99% 95% Table 2 OCAv3 Performance Characteristic Results for all Variant Types Table 1 LOD Performance for INDEL Called by Ion ReporterTM (IR) Using a Cell Line Dilution Series Dilution Series 1 2 3 4 5 6 7 6.1% Replicate 1 Replicate 2 EGFR p.Glu746_Ala750del Allele Frequency 87.0% 22.0% 89.0% 20.0% 5.7% 3.8% 3.2% NDND 4.9% 3.5% 1.3% ND A B C
