Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrating T cell repertoire
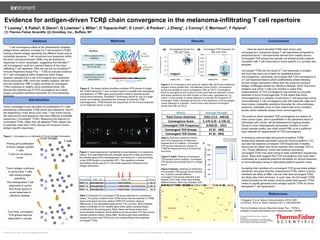
- 1. For Research Use Only. Not for use in diagnostic procedures. Thermo Fisher Scientific • 5781 Van Allen Way • Carlsbad, CA 92008 • thermofisher.com T Looney1, S Pabla2, S Glenn2, G Lowman1, L Miller1, D Topacio-Hall1, E Linch1, A Pankov1, J Zheng1, J Conroy2, C Morrison2, F Hyland1. (1) Thermo Fisher Scientific (2) OmniSeq, Inc., Buffalo, NY Evidence for antigen-driven TCRβ chain convergence in the melanoma-infiltrating T cell repertoire Abstract T cell convergence refers to the phenomenon whereby antigen-driven selection enriches for T cell receptors (TCRs) having a shared antigen specificity but different amino acid or nucleotide sequence. T cell recruitment and expansion within the tumor microenvironment (TME) may be directed by responses to tumor neoantigen, suggesting that elevated T cell convergence could be a general feature of the tumor infiltrating T cell repertoire. Here we use the Ion AmpliSeq™ Immune Repertoire Assay Plus – TCRβ to evaluate evidence for T cell convergence within melanoma tumor biopsy research samples from a set of 63 subjects plus peripheral blood leukocytes (PBL) from four healthy subjects. We find that the melanoma TME is highly enriched for convergent TCRs compared to healthy donor peripheral blood. We discuss the potential use of TCR convergence as a liquid- biopsy compatible predictive biomarker for immunotherapy response. For Research Use Only. Not for use in diagnostic procedures. © 2018 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Conclusions 1.Ruggiero, E et al. Nature Communications (2015). 8081 2.Emerson, RO et al. Nature Genetics (2017). 49(5):659-665 The Ion Ampliseq Immune Repertoire Assay Plus – TCRβ is available via www.thermofisher.com Catalog Number: A35386 Methods References Introduction Results Figure 4. A) Convergent clone score for healthy PBL (N=4) and melanoma biopsies having greater than 100 detected clones (N=63). Convergence scores are elevated in tumor compared to PBL (p<1E-7). Convergence score is calculated as the number of clone pairs that are identical in amino acid space divided by the total number of clone pairs, calculated as nCr2, where n is the number of clones detected in a sample. B) Frequency of convergent TCRs, calculated as the sum of the frequency of all convergent clones detected in a sample. Tumors have more frequent convergent clones than PBL (p<1E-4). Here we report elevated TCRβ chain amino acid convergence in melanoma biopsy T cell repertoires compared to peripheral blood T cell repertoires derived from healthy donors. Convergent TCR groups are typically not shared across subjects, consistent with T cell responses to tumor-specific (i.e. private) neo- epitopes. Convergent TCRs are the result of T cell responses to antigen, and thus may serve as a metric for quantifying tumor immunogenicity. Importantly, we propose that TCR convergence is a T cell repertoire feature which preferentially arises following chronic antigen stimulation rather than the acute but transient antigen challenges elicited by infectious disease. Given that tumor antigens may prime T cells over months or years time, measurements of TCR convergence may provide an accurate estimate of tumor immunogenicity. To the extent that tumor immunogenicity is a predictor of anti-tumor T cell responses during immunotherapy, T cell convergence may hold particular utility as a liquid biopsy compatible predictive biomarker for immunotherapy response, potentially more so than metrics like tumor mutation burden which indirectly assess tumor immunogenicity. The extent to which elevated TCR convergence is a feature of other cancer types, and is quantifiable in the peripheral blood of individuals with cancer, will be addressed by ongoing studies. Owing to tumor heterogeneity and greater variation in tumor biopsy sample quality, one might predict PBL to be a preferred input material for measurement of TCR convergence. In reviewing commercially-derived and academic TCRβ sequencing datasets generated using the Illumina platform (1,2), we note that baseline convergent TCR frequencies in healthy donors are far higher than those reported here (average 15% vs .2%, 75-fold difference). Given that artifacts resembling convergent TCRs may arise owing to base substitution sequencing errors, these data suggest that TCR convergence may have been overlooked as a potential predictive biomarker for clinical response to immunotherapy owing to obfuscating platform-specific noise. Accepting that members of a convergent TCR group share antigen specificity, and given that the unsequenced TCRα chains of group members are likely to differ, one can infer that convergent TCRs are likely beta chain dominant. In such case, the full length TCRβ chains provided by this assay could be paired with generic TCRα chains to rapidly generate tumor antigen specific TCRs for future therapeutic T cell applications. Healthy PBL Tumor -9 -8 -7 -6 -5 Convergence Scores for PBL and Tumor Log10ConvergenceScore Healthy PBL Tumor 0.00 0.01 0.02 0.03 0.04 0.05 0.06 Convergent TCR Frequency for PBL and Tumor ConvergentTCRFrequency Number of individuals sharing convergent TCRB group 0 200 400 600 800 1000 1200 1400 1 2 3 4 5 6 7 8 10 CDR loops T Cell Receptor HLA presenting peptide Variable Diversity Joining Constant N1 N2 A. Adult IGH orTCRBeta chain rearrangement In adult B andT cells, the process ofVDJ rearrangement very often involves exonucleotide chewback ofVDJ genes and the addition of non-templated bases, forming N1 and N2 regions in the B cell receptor heavy chain CDR3 and theT cell receptor Beta chain CDR3. These processes vastly increase IGH andTCRB CDR3 diversity. Variable Diversity Joining Constant B. Fetal IGH orTCRBeta chain rearrangement In the fetus, the process ofVDJ rearrangement often occurs without exonucleotide chewback ofVDJ genes and addition of non-templated bases, resulting in a restricted IGH andTCRB CDR3 repertoire that is distinct from the adult repertoire. These structural differences can be used to distinguish fetal B andT cell CDR3 receptors from maternal B andT cell CDR3 receptors in cell free DNA present in maternal peripheral blood. In this way, fetal B andT cell health and development may be monitored in a non-invasive manner. Figure 1. Structural differences between fetal and adult B andT cell receptors ~330bp amplicon 1B Sequencing in multiplex on S5 via 530 chip FR1-C multiplex PCR ( AmpliSeq) Ion Reporter analysis of repertoire features and polymorphism Workflow Figure 2. The assay utilizes AmpliSeq multiplex PCR primers to target the TCRβ Framework 1 and Constant regions to enable both clonotyping and detection of TRBV gene polymorphism linked to adverse events during immunotherapy. AmpliSeq™ technology allows for use of a large primer set enabling comprehensive coverage of potential TCRβ rearrangements. TCRβ libraries are sequenced via the S5 and analyzed on Ion Reporter server or cloud. Tumor Antigen T cells Variable CDR3 AA CDR3 NT Freq TRBV7-8 ASSLGQAYEQY GCCAGCAGCTTAGGTCAGGCATACGAGCAGTAC 1.8E-3 TRBV7-8 ASSLGQAYEQY GCCAGCAGCTTGGGACAGGCCTACGAGCAGTAC 4.8E-4 TRBV7-8 ASSLGQAYEQY GCCAGCAGCTTAGGGCAGGCCTACGAGCAGTAC 1E-4 ● 0 20 40 60 80 TCRB V−gene usage and number of clones for TCR−005−RS−02738603_RNA_OIRRA−7_RNA_v1_20170517212600606 CDR3length(nt) 1 23 46 Number of Clones 201100 readsClone Shannon Diversity: 6.6392 Clone Evenness: 0.6278 1525 clones Mean CDR3 NT length: 35.4133 +/− 5.3053 V−gene Shannon Diversity: 3.9098 V−CDR3 Frequency 0.10 0.05 ●0.01 ●0.005 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● TRBV1 TRBV2 TRBV3−1 TRBV4−1 TRBV5−1 TRBV6−1 TRBV7−1 TRBV4−2 TRBV6−2 TRBV3−2 TRBV4−3 TRBV6−3 TRBV7−2 TRBV6−4 TRBV7−3 TRBV5−3 TRBV9 TRBV10−1 TRBV11−1 TRBV12−1 TRBV10−2 TRBV11−2 TRBV12−2 TRBV6−5 TRBV7−4 TRBV5−4 TRBV6−6 TRBV5−5 TRBV6−7 TRBV7−6 TRBV5−6 TRBV6−8 TRBV7−7 TRBV5−7 TRBV6−9 TRBV7−8 TRBV5−8 TRBV7−9 TRBV13 TRBV10−3 TRBV11−3 TRBV12−3 TRBV12−4 TRBV12−5 TRBV14 TRBV15 TRBV16 TRBV17 TRBV18 TRBV19 TRBV20−1 TRBV21−1 TRBV23−1 TRBV24−1 TRBV25−1 TRBV26 TRBV27 TRBV28 TRBV29−1 TRBV30 Figure 3. Spectratyping plot highlighting clones detected in a melanoma biopsy. VDJ rearrangements are arranged along the X-axis according to the variable gene of the rearrangement, and along the Y- axis according to the CDR3 length in nucleotides (NT). This repertoire includes dominating clones typical of T cell clonal expansion following tumor antigen stimulation. Table 1. Example of a convergent TCR group detected in a melanoma biopsy. This group contains three TCRβ clones that are identical in TCRβ amino acid space but have distinct CDR3 NT junctions owing to differences in non-templated bases at the V-D-J junction. Blue indicates bases contributed by the variable gene while yellow indicates bases contributed by the joining gene. Black bases are those arising from exonucleotide chewback and N-additions by the enzyme TdT. Red arrows indicate positions where clones differ. Illumina-style base substitution sequencing errors and PCR errors can create artifacts that resemble convergent TCRs. Metric Average (Range) Total Clones Detected 5262 (113 - 44610) Convergence Score 5.17E-6 (0 - 4.19E-5) Convergent TCR Frequency 0.024 (0 - .231) Convergent TCR Groups 22 (0 - 183) Convergent TCR Clones 47 (0 - 399) 4A 4B Table 5. Summary repertoire metrics from TCR sequencing of melanoma biopsies from 63 subjects. Convergent TCR groups indicates the number of TCRβ AA sequences found in more than one clone. Figure 5 (right). Sharing of convergent TCR groups across subjects. Convergent TCR groups are typically found in a single donor. Figure 6 (below). Heatmap of distribution of convergent TCR groups across sample set. Columns indicate different convergent TCR groups detected in the dataset (1341 total), while rows indicate different donors. Red color indicates TCR group is abundant in a subject. Tumor neoantigens may stimulate the proliferation of T cells possessing a stereotyped TCRβ amino acid sequence. Due to the degeneracy of the amino acid code, T cell clones having the same amino acid sequence may have different nucleotide sequences (“convergent” TCRs). Measuring the features of convergent TCRs, rather than all detected TCRs, allows one to eliminate noise from TCRs that are not involved in tumor antigen specific responses. Figure 1. Convergence is a Hallmark of Chronic Antigen Stimulation Priming and proliferation of tumor antigen specific T cells. T cells do not destroy tumor. Tumor antigen continues to prime naive T cells with shared antigen specificity. This distinguishes T cell responses to cancer from those typical of acute responses to infectious disease.. Over time, convergent TCR groups become detectable in sample. NumberofconvergentTCRBgroups 5 Number of samples having TCR group Convergence Scores For PBL and Tumor Convergent TCR Frequency for PBL and Tumor
