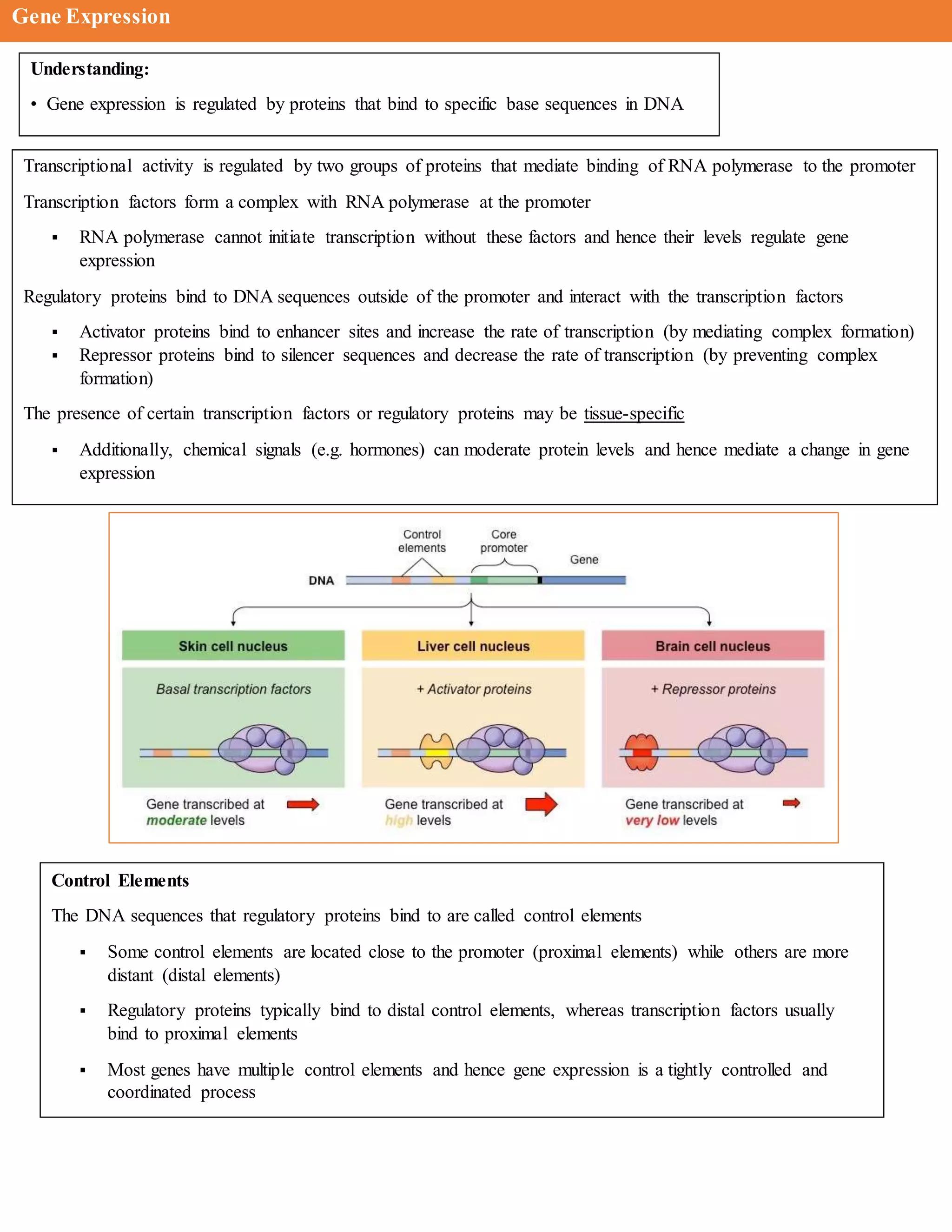
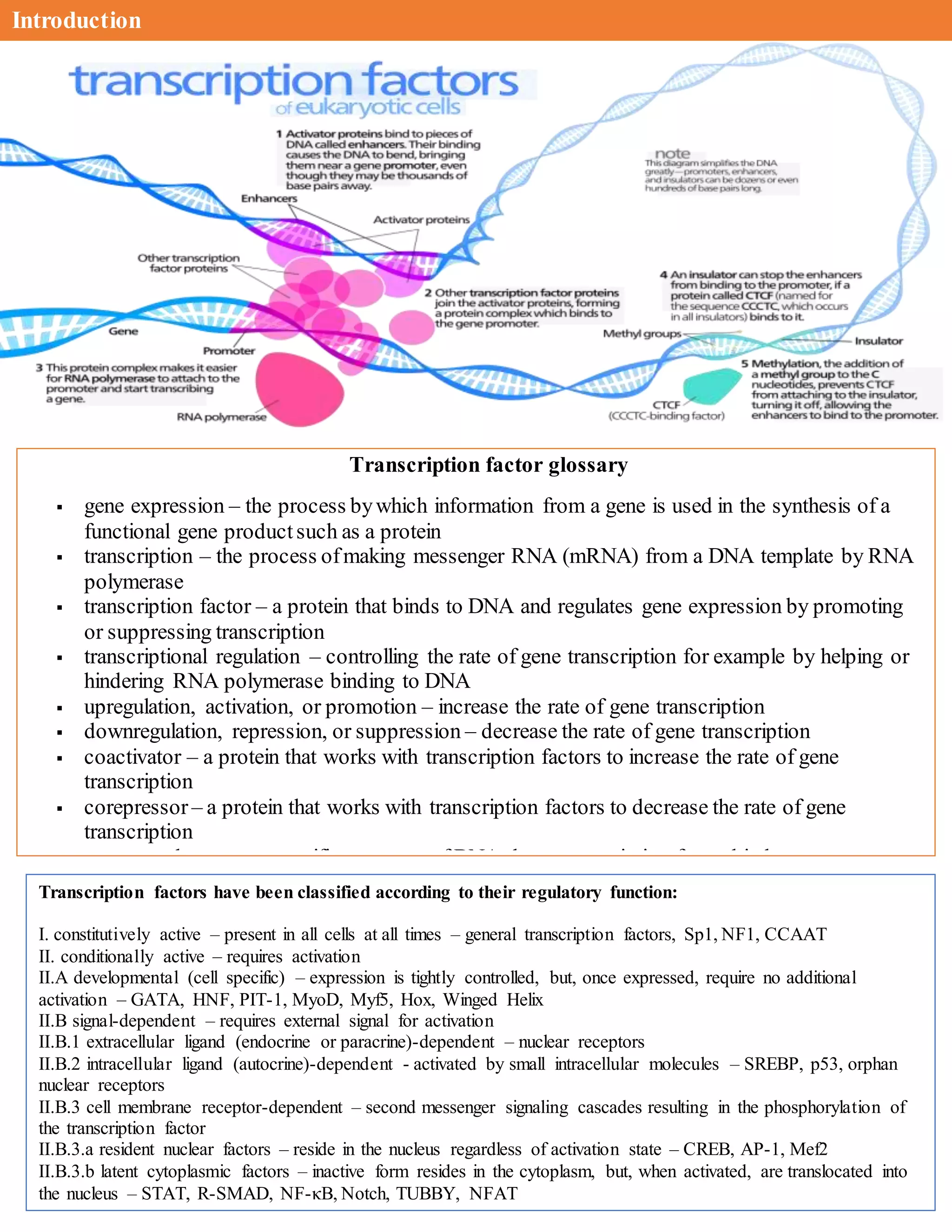
The document discusses the role of transcription factors in gene expression, detailing their classifications, functions, and mechanisms of action, including both enhancers and repressors. It also highlights the significance of transcriptional regulation in disease, outlining specific mutations in transcription factors that are linked to various syndromes and cancers. Furthermore, the document presents key concepts related to RNA polymerase, promoters, and the intricacies of transcription initiation.

![So, what is activator?
Activator (genetics)
A transcriptional activator is a protein (transcription factor) that increases gene
transcription of a gene or set of genes. Most activators are DNA-binding proteins that bind
to enhancers or promoter-proximal elements.
Most activators function by binding sequence-specifically to a DNA site located in or near
a promoter and making protein–protein interactions with the general transcription
machinery (RNA polymerase and general transcription factors), thereby facilitating the
binding of the general transcription machinery to the promoter. The DNA site bound by
the activator is referred to as an "activator site". The part of the activator that makes
protein–protein interactions with the general transcription machinery is referred to as an
"activating region". The part of the general transcription machinery that makes protein–
protein interactions with the activator is referred to as an "activation target".
The catabolite activator protein (CAP; also known as cAMP receptor protein, CRP)activates
transcription at the lac operon of the bacterium Escherichia coli.[1] Cyclic adenosine monophosphate
(cAMP) is produced during glucose starvation, binds to CAP, causes a conformational change that
allows CAP to bind to a DNA site located adjacent to the lac promoter. CAP then makes a direct
protein–protein interaction with RNA polymerase that recruits RNA polymerase to the lac promoter.](https://image.slidesharecdn.com/molecular-200723084711/75/Activation-of-gene-expression-by-transcription-factors-5-2048.jpg)