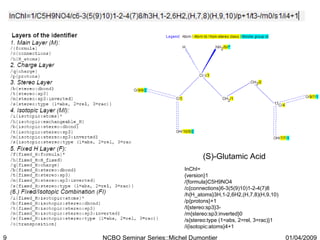
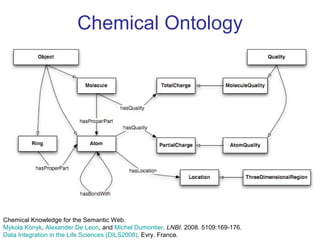
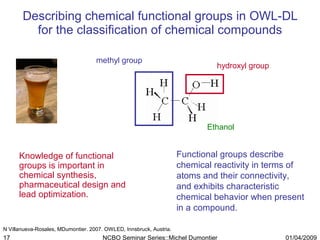
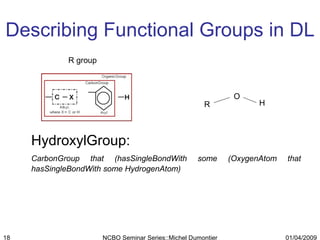
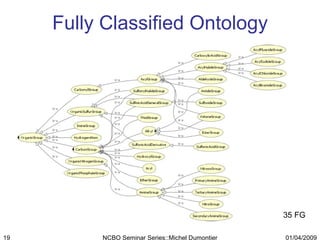
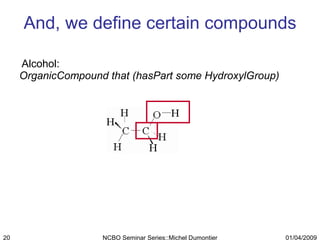
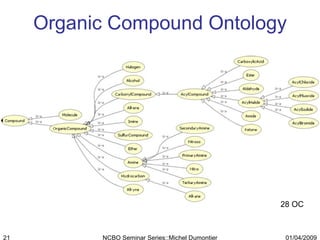
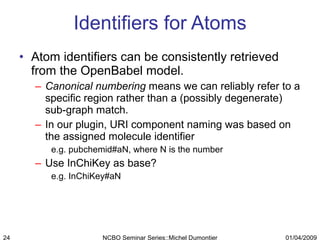
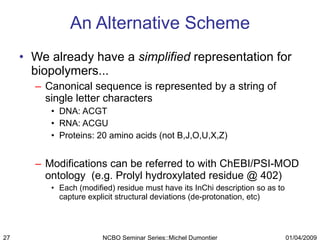
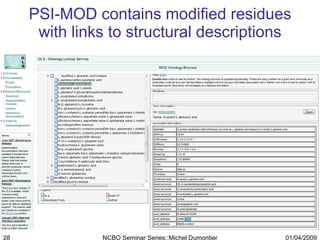
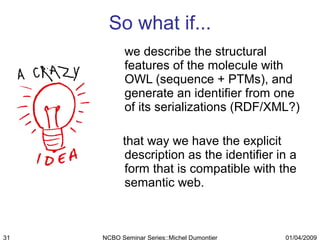
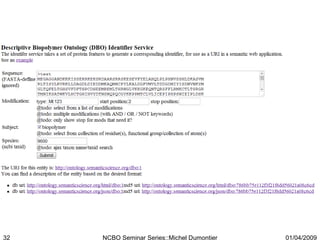
The document discusses the need for precise identifiers for biochemical entities based on their structural characteristics to improve the granularity of biochemical knowledge representation. It introduces issues with existing databases and proposes methodologies for generating unique identifiers for biopolymers and their components. The aim is to establish a consistent naming convention that links biochemical information more effectively across various contexts.







([C@H](O)([C@@H](O)([C@@H]1(O)))))(CO) 79025 IUPAC InChI=1/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3-,4+,5-,6+/m1/s1 InCHI α -D-Glucose 6-(hydroxymethyl)oxane-2,3,4,5-tetrol OR (2R,3R,4S,5R,6R)-6 -(hydroxymethyl)tetrahydro -2H-pyran-2,3,4,5-tetraol SMILES](https://image.slidesharecdn.com/20090401ncbostructuralidentity-090401100221-phpapp01/85/Accurate-biochemical-knowledge-starting-with-precise-structure-based-criteria-for-molecular-identity-13-320.jpg)






![While we’re at it, we could extend our expressive capability to match that of OWL: Specification Exactly mod1@pos X Only mod1@posX Minimum : At least [email_address] Combination: mod1@posX AND mod2@posY, X != Y Possibilities/Uncertainty: (mod1 OR mod2) @posX Exclusion : not mod1 @ posX 01/04/2009 NCBO Seminar Series::Michel Dumontier](https://image.slidesharecdn.com/20090401ncbostructuralidentity-090401100221-phpapp01/85/Accurate-biochemical-knowledge-starting-with-precise-structure-based-criteria-for-molecular-identity-30-320.jpg)



![dumontierlab.com [email_address] Special thanks to PhD Student Leonid Chepelev for insightful discussions semanticscience.org 01/04/2009 NCBO Seminar Series::Michel Dumontier](https://image.slidesharecdn.com/20090401ncbostructuralidentity-090401100221-phpapp01/85/Accurate-biochemical-knowledge-starting-with-precise-structure-based-criteria-for-molecular-identity-37-320.jpg)