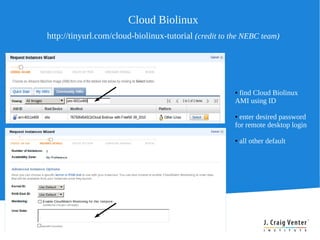
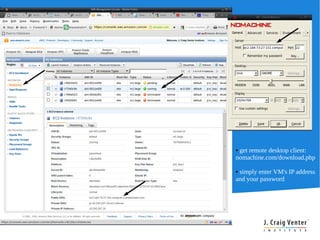
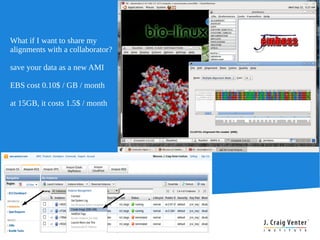
Cloud BioLinux provides pre-configured virtual machines on Amazon EC2 with over 100 bioinformatics tools for genomic analysis. It allows small labs and researchers to perform large-scale genomic computations in the cloud without extensive hardware, expertise, or time to install software locally. The Cloud BioLinux virtual machine images can be run on EC2 or other cloud platforms like Eucalyptus. The project is open source and community-driven to expand available tools and collaboration capabilities.