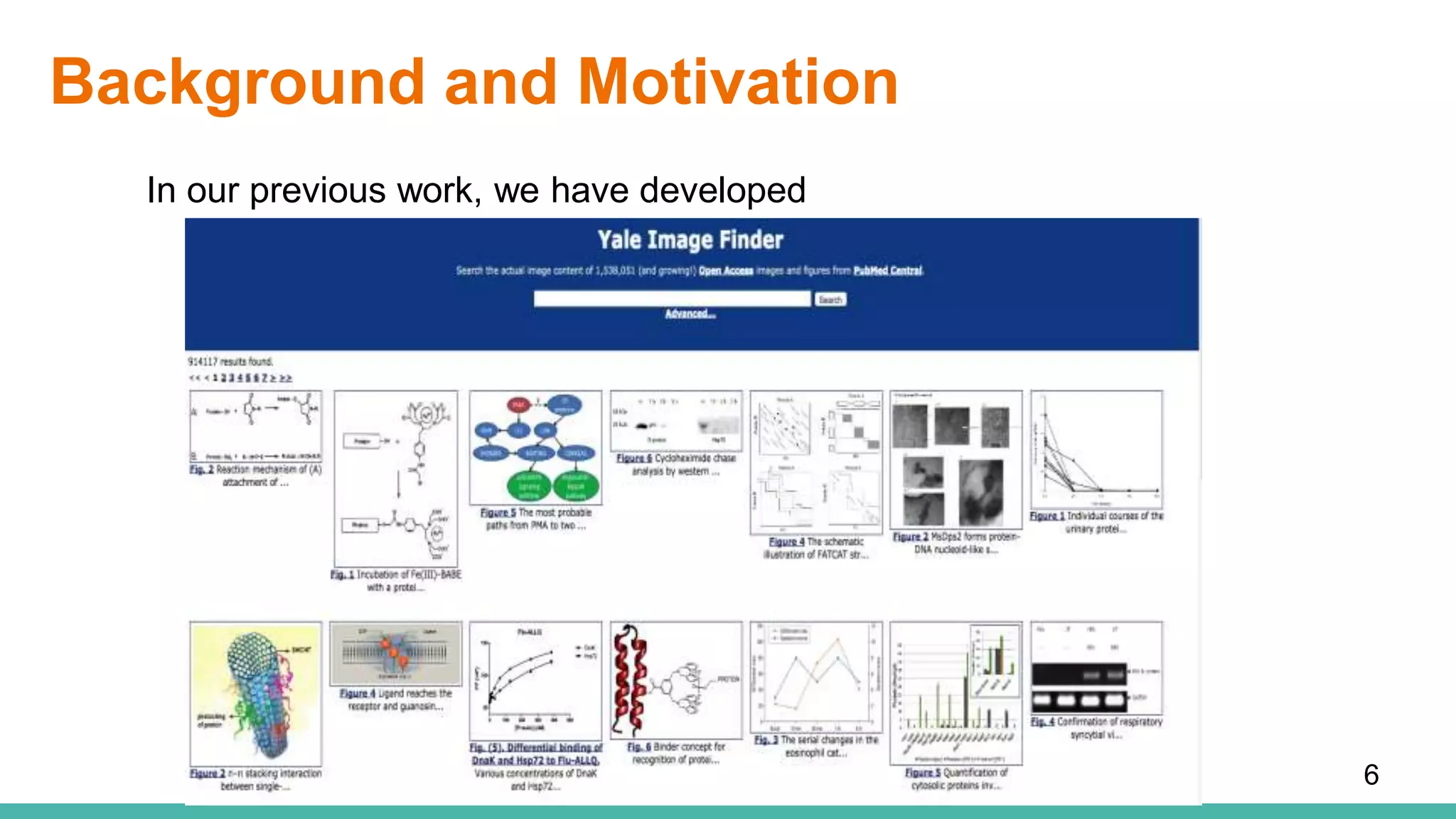
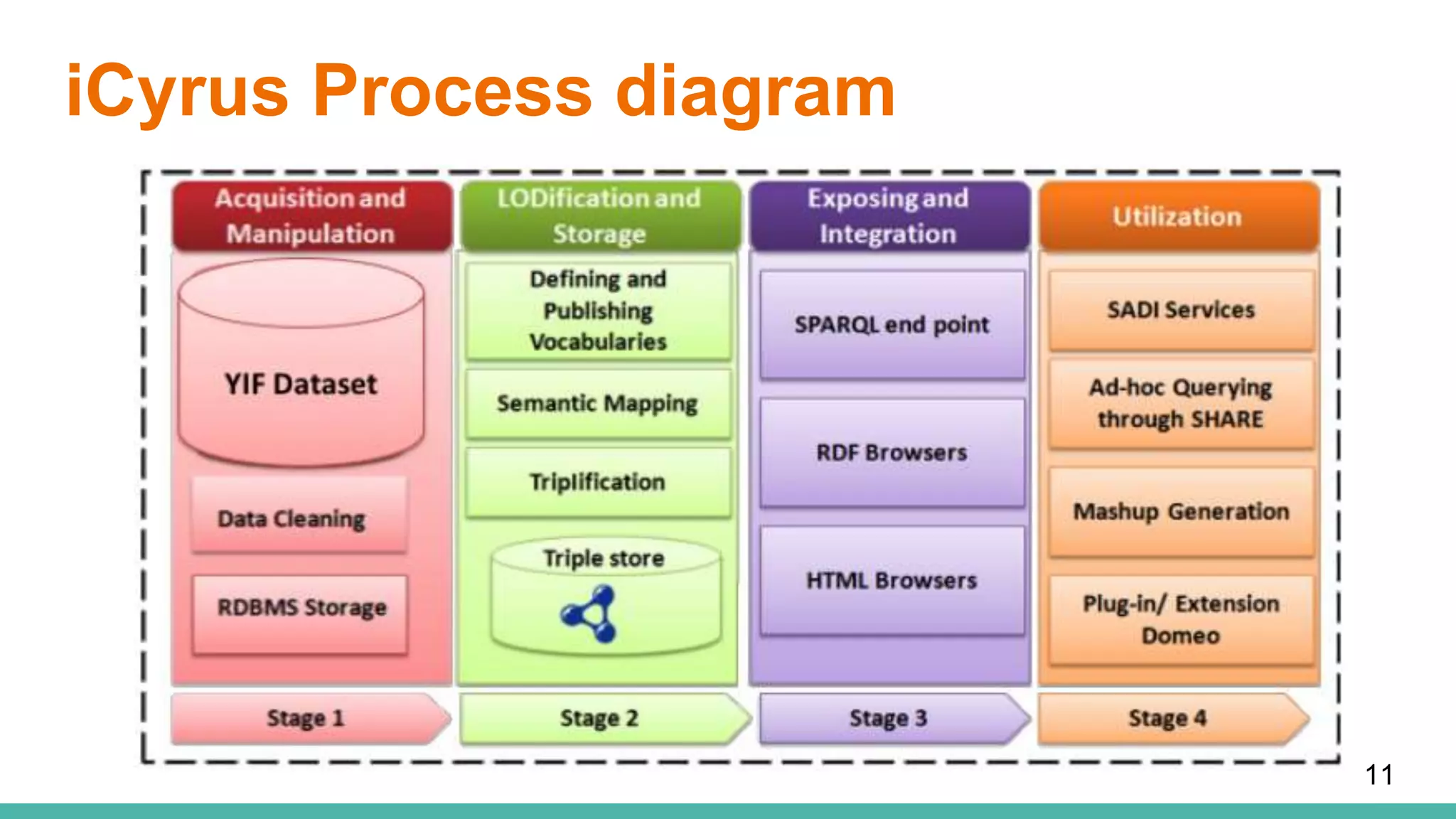
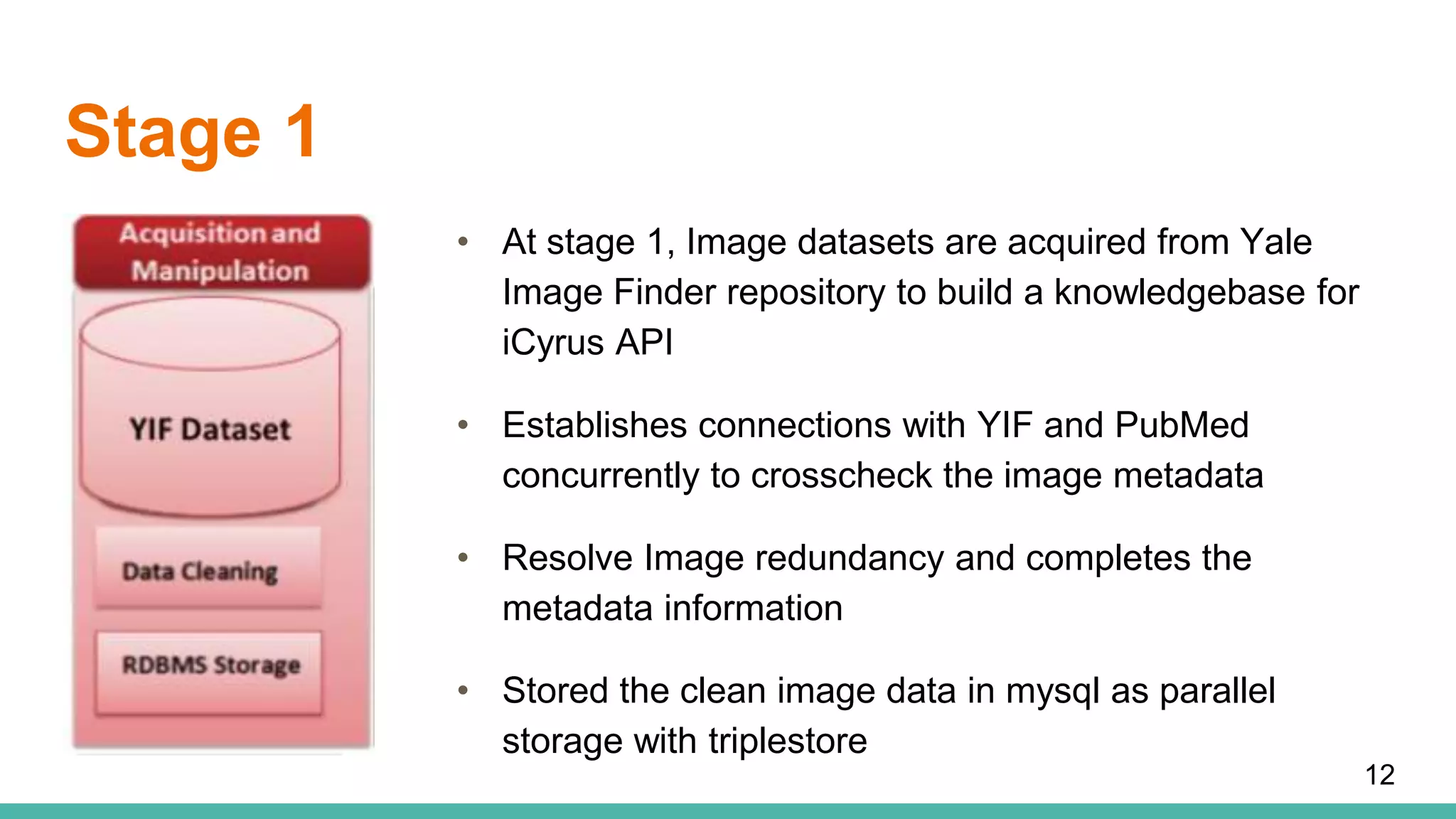
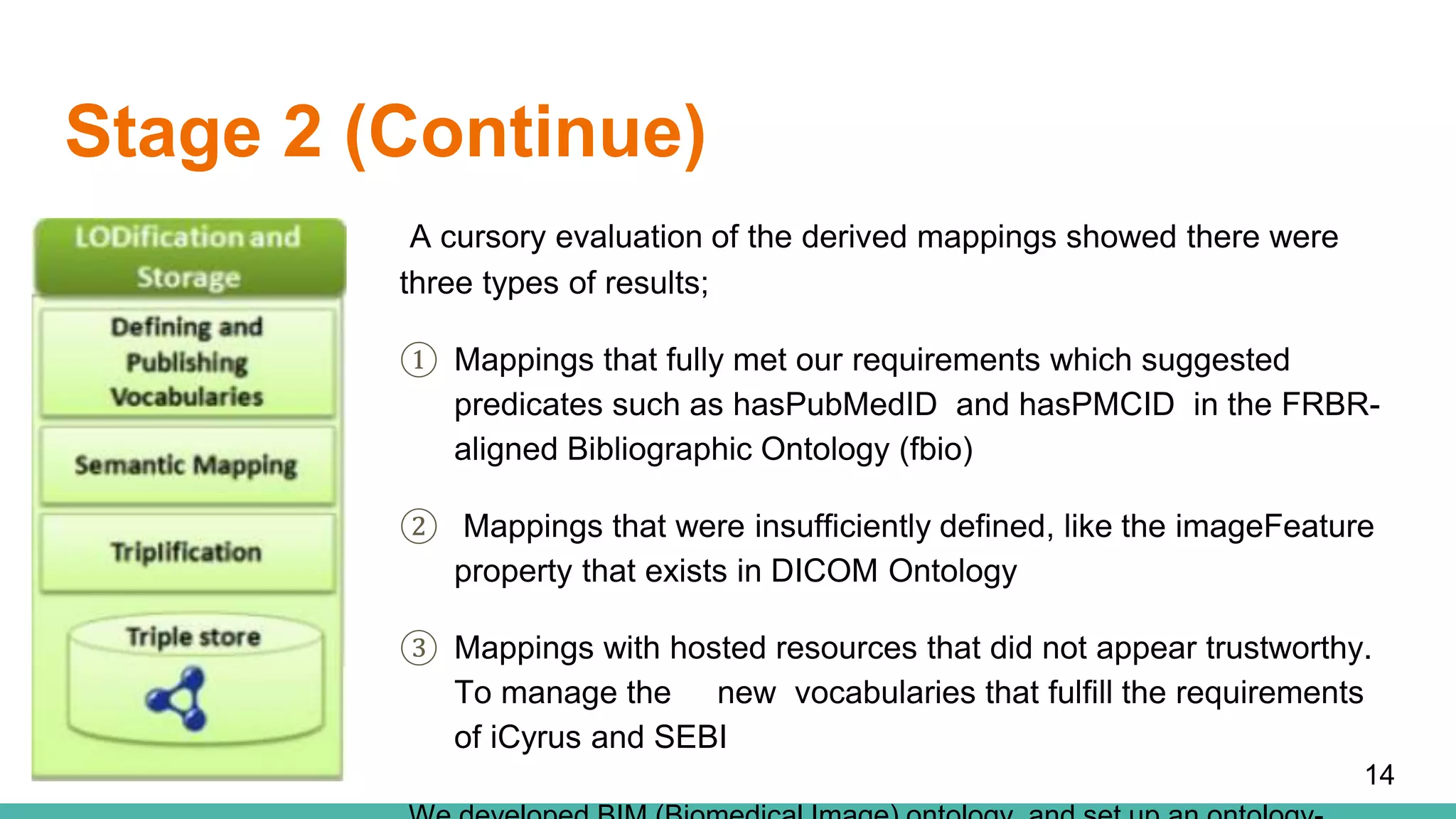
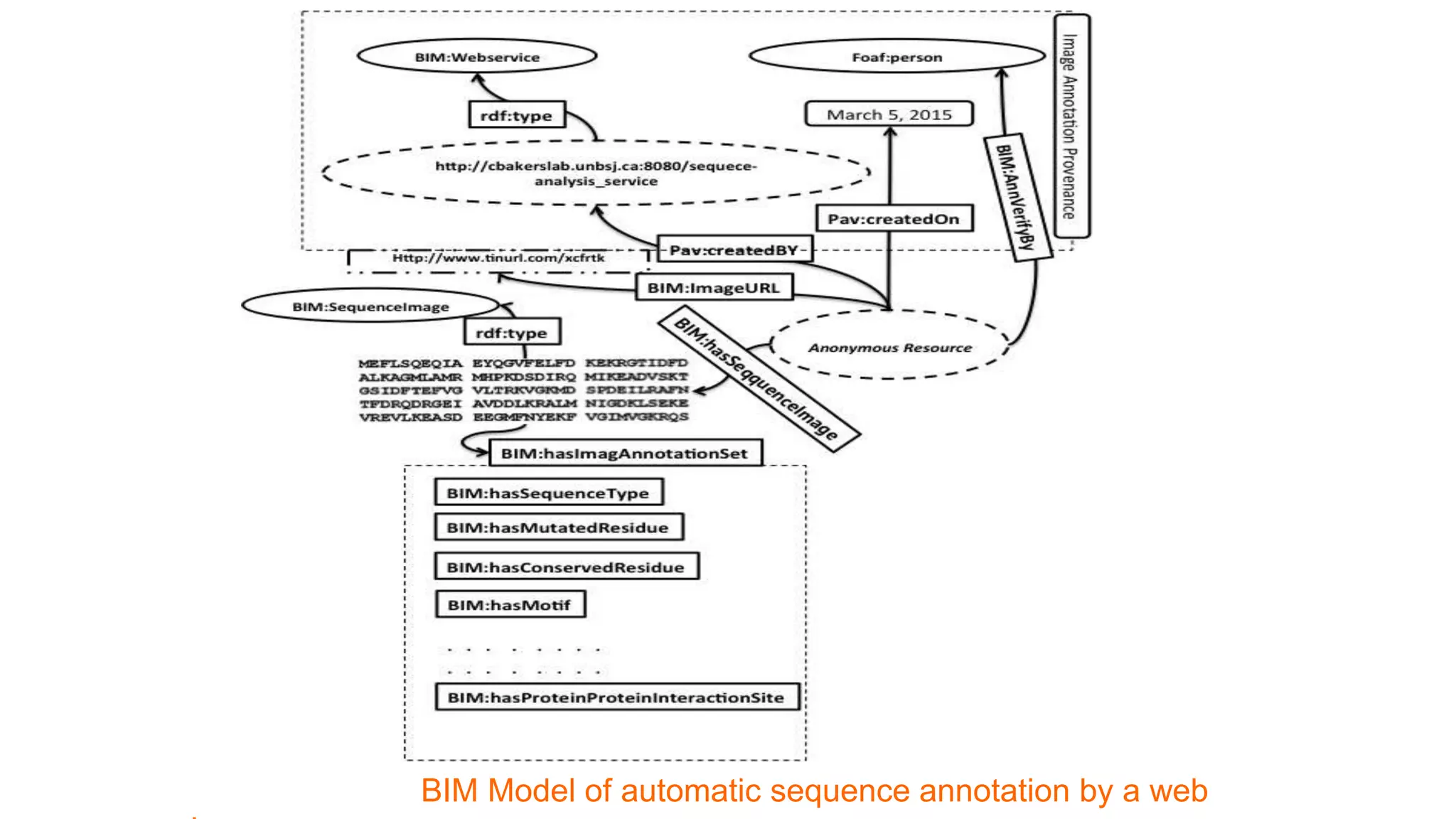
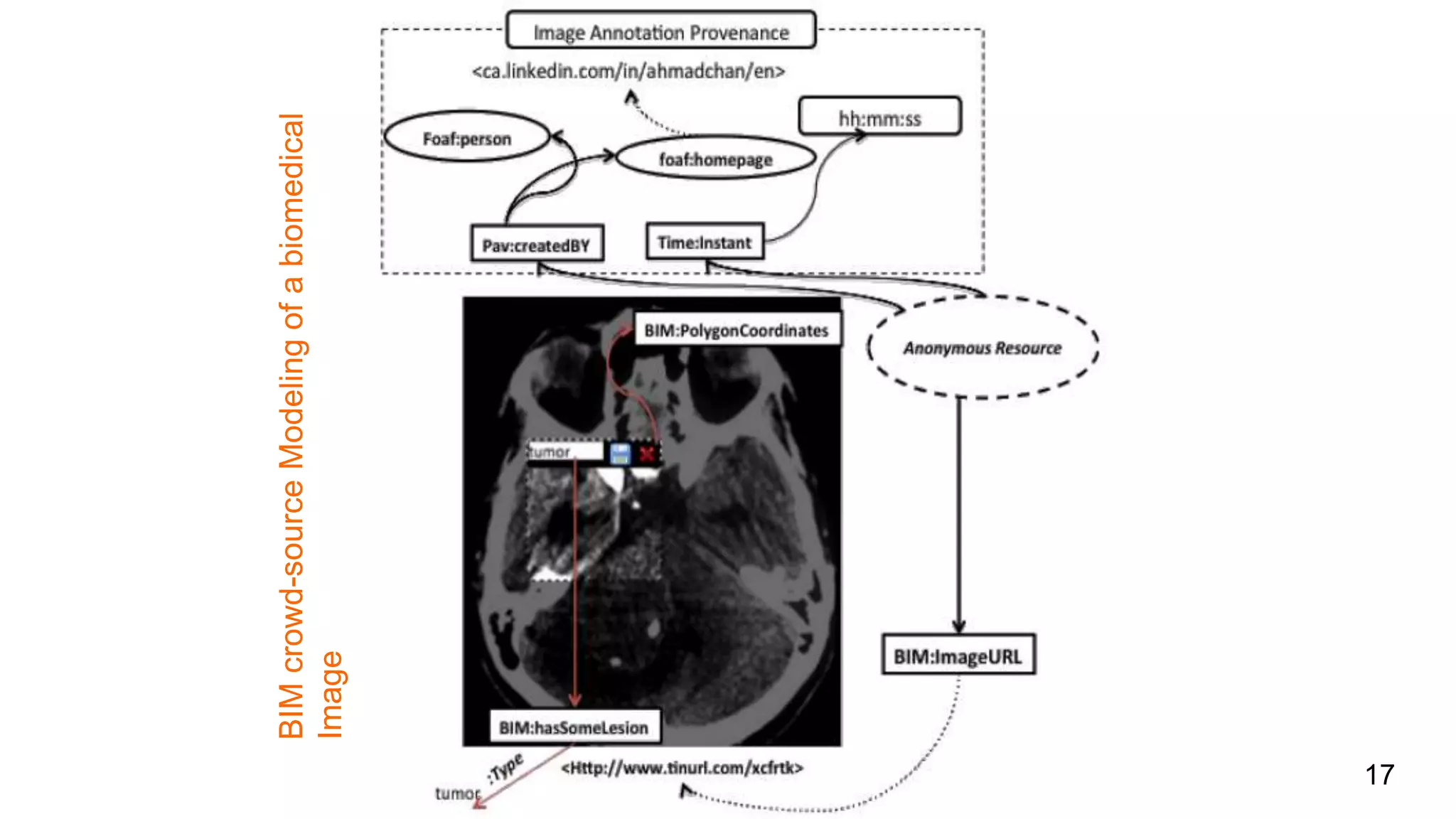
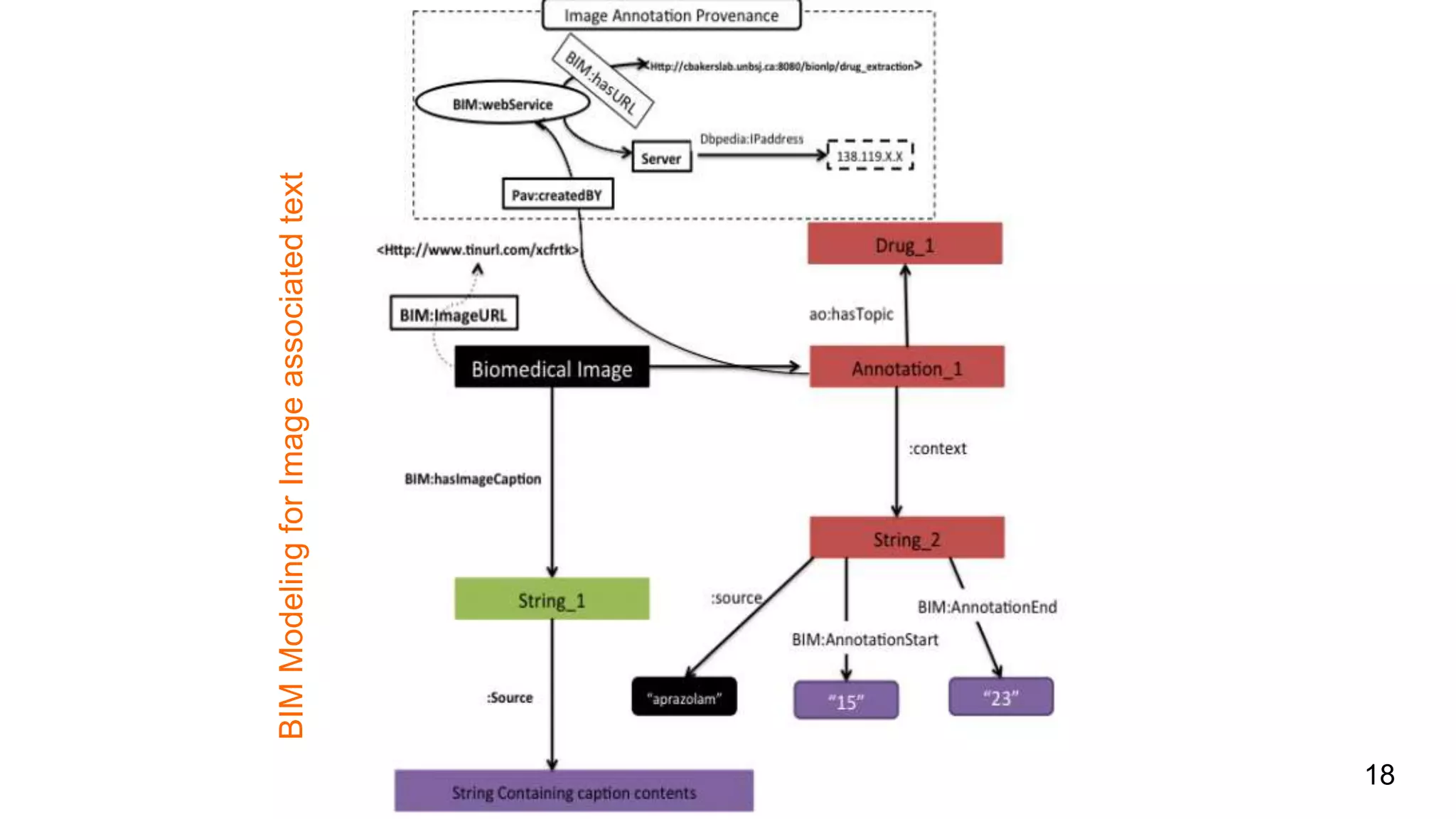
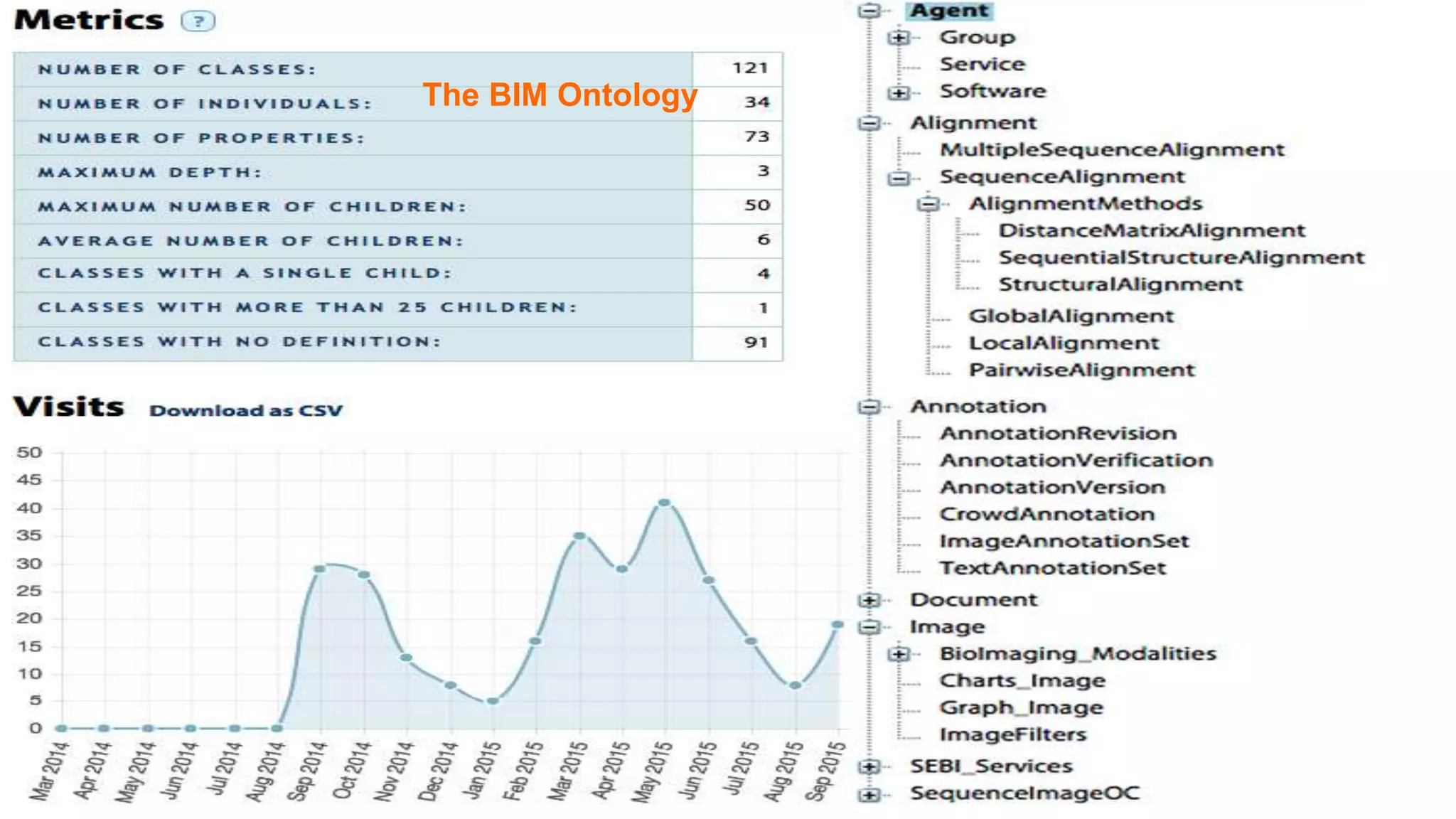
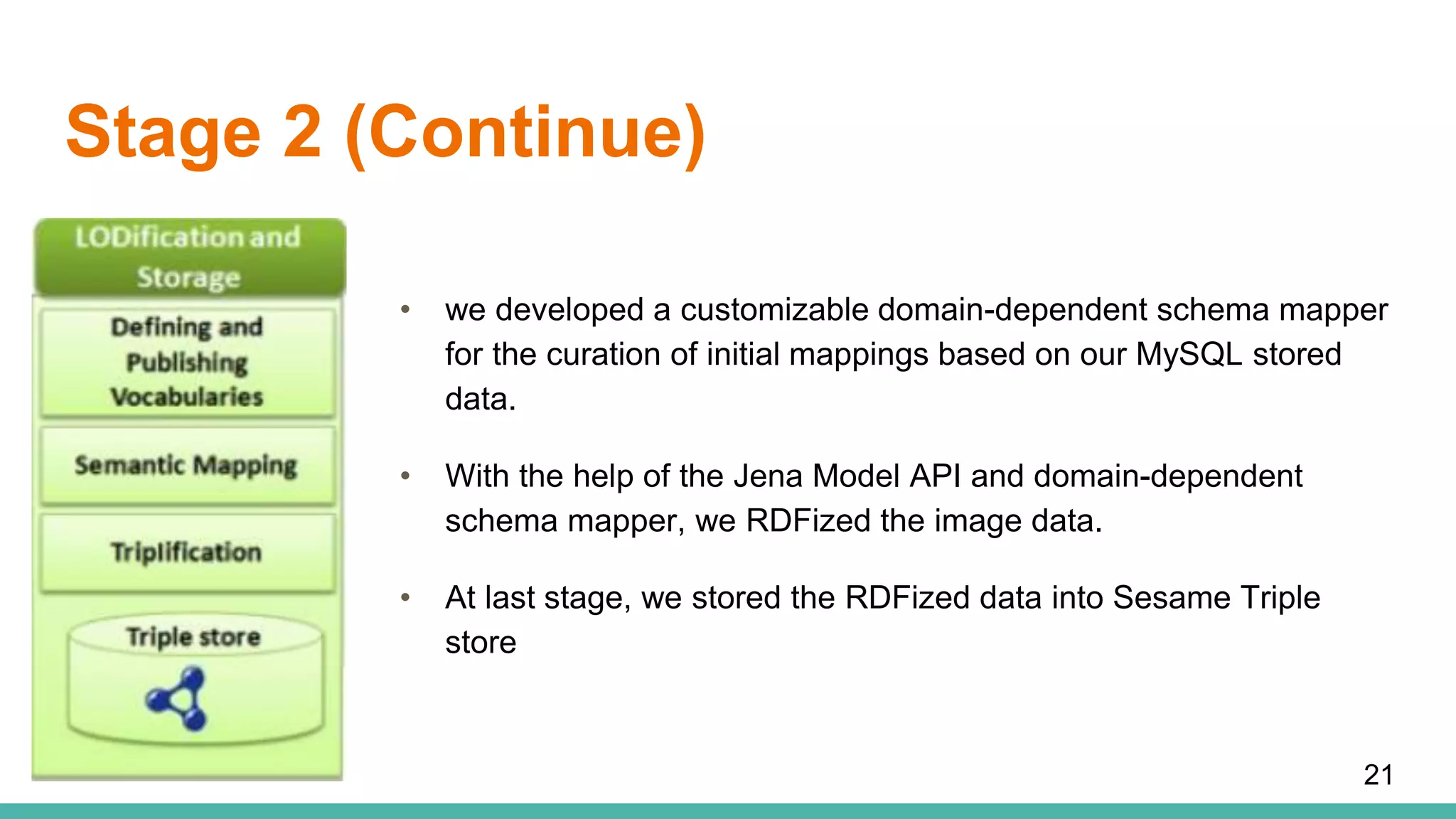
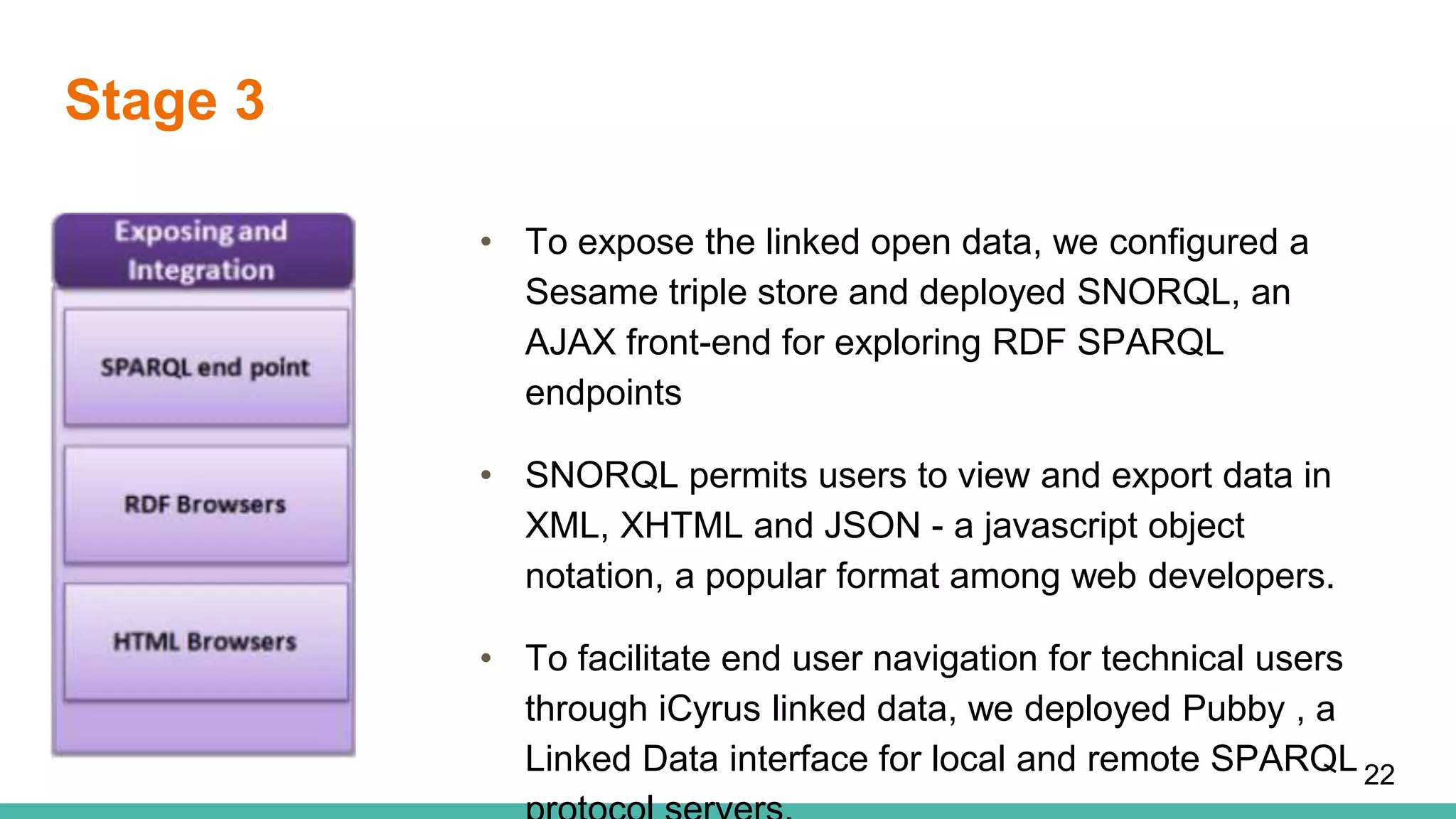
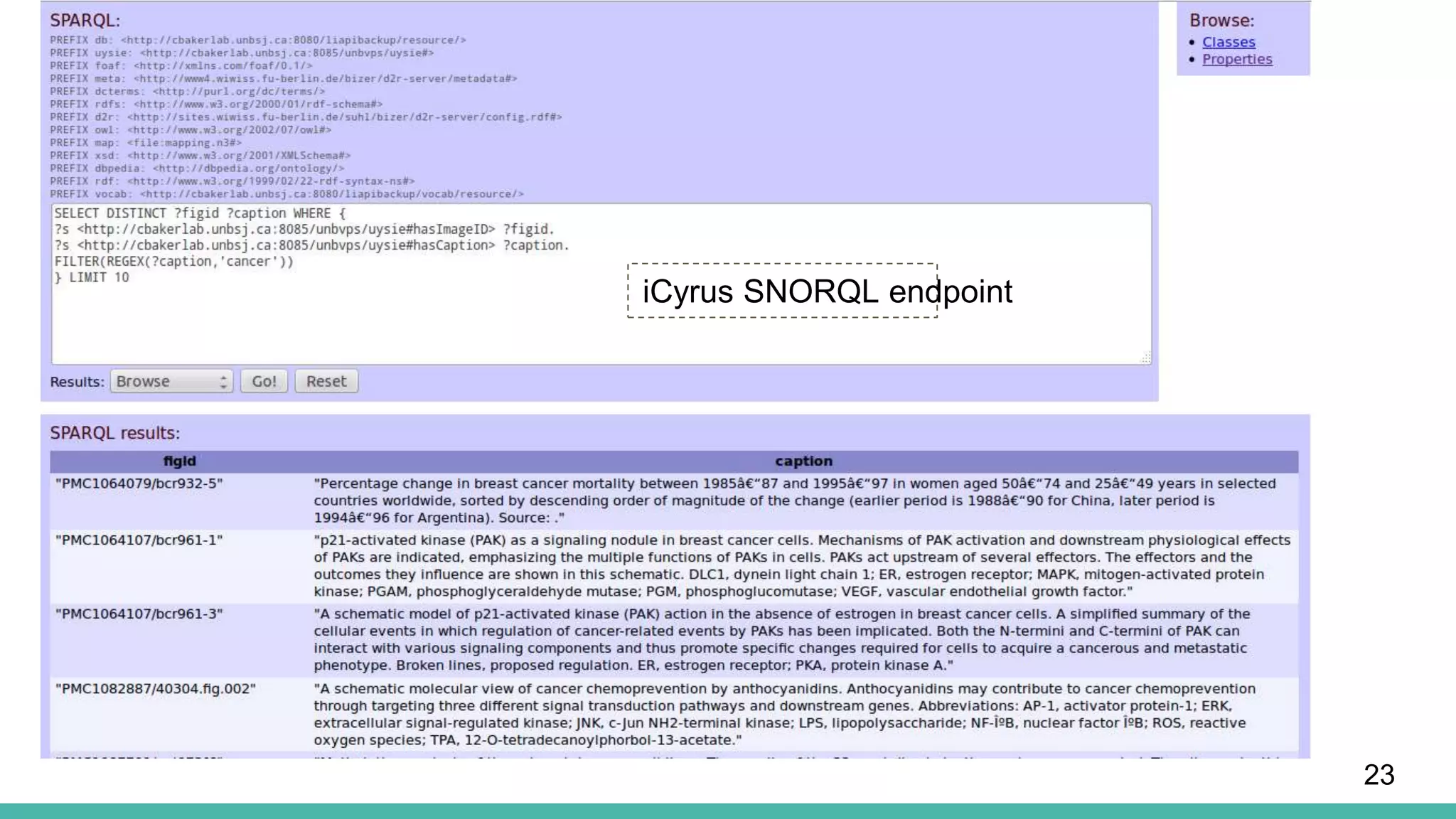
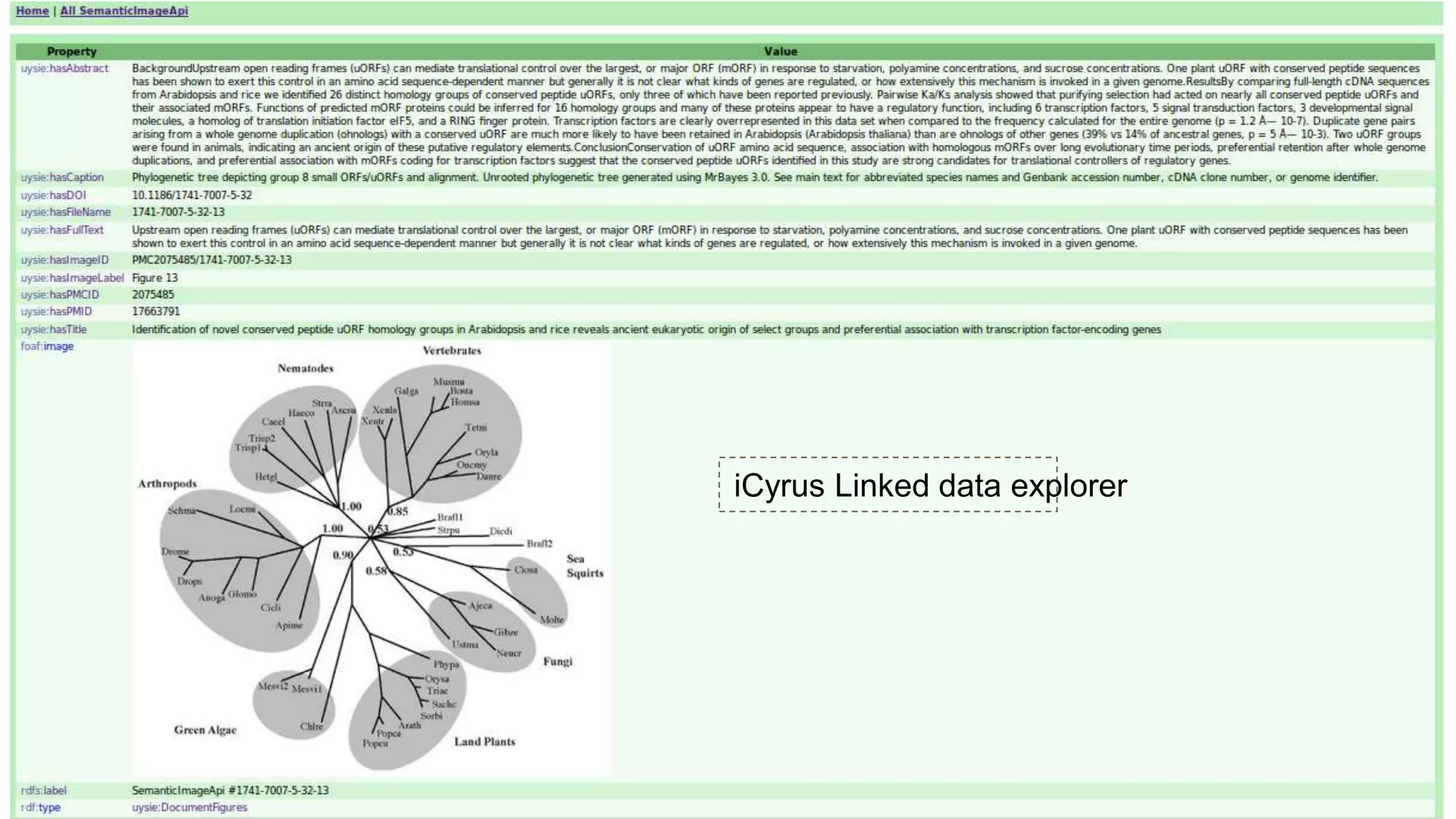
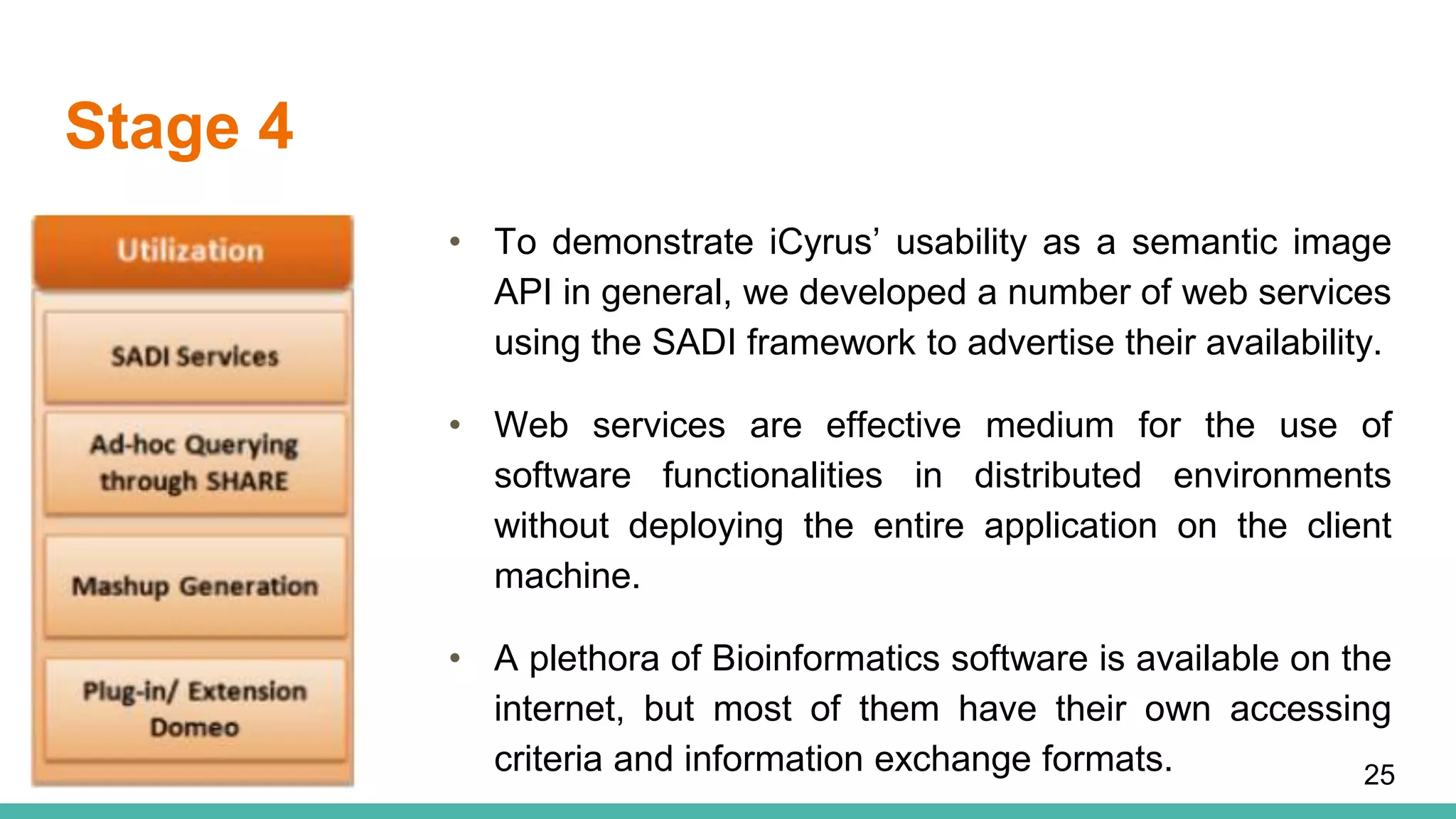
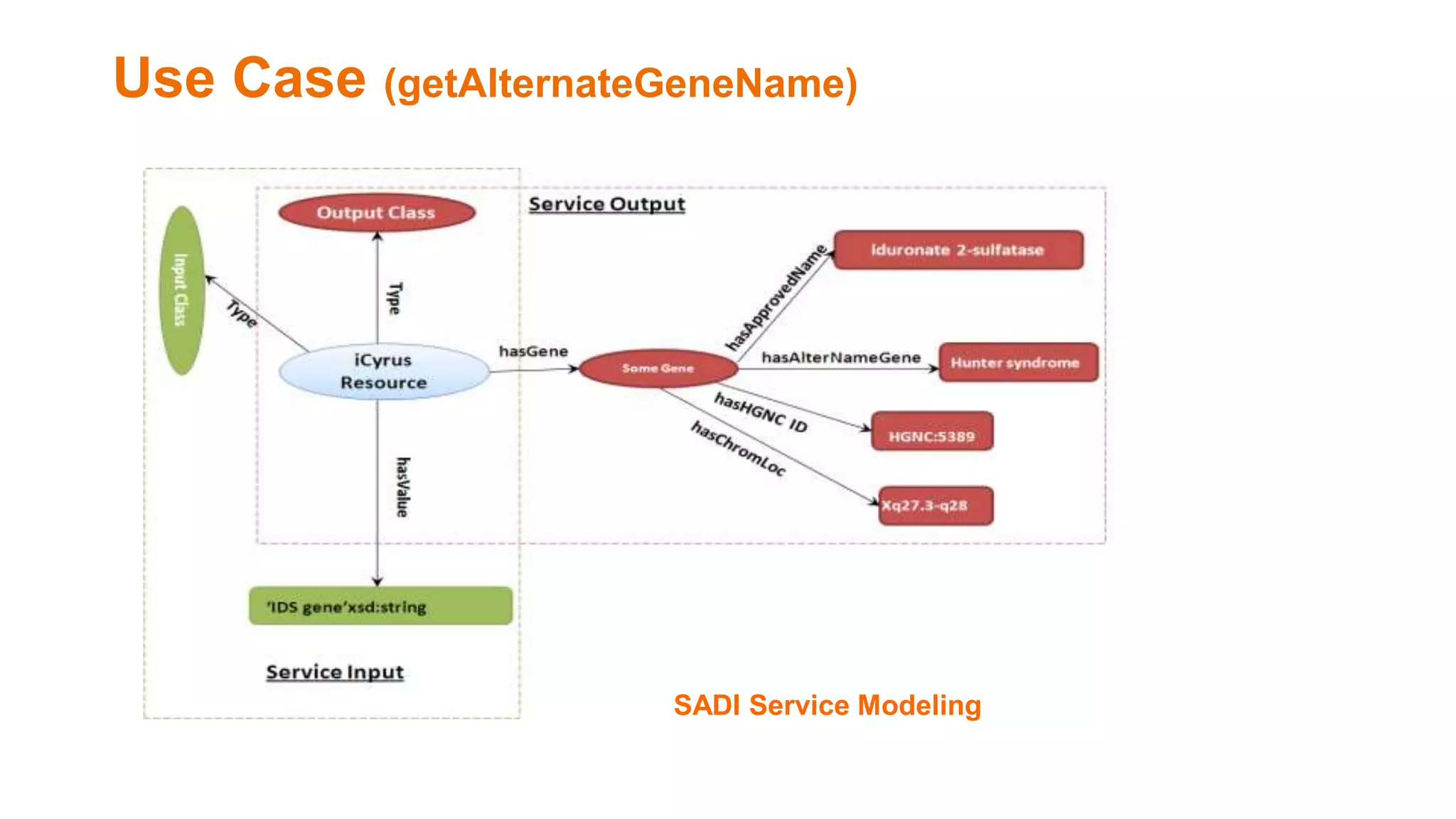
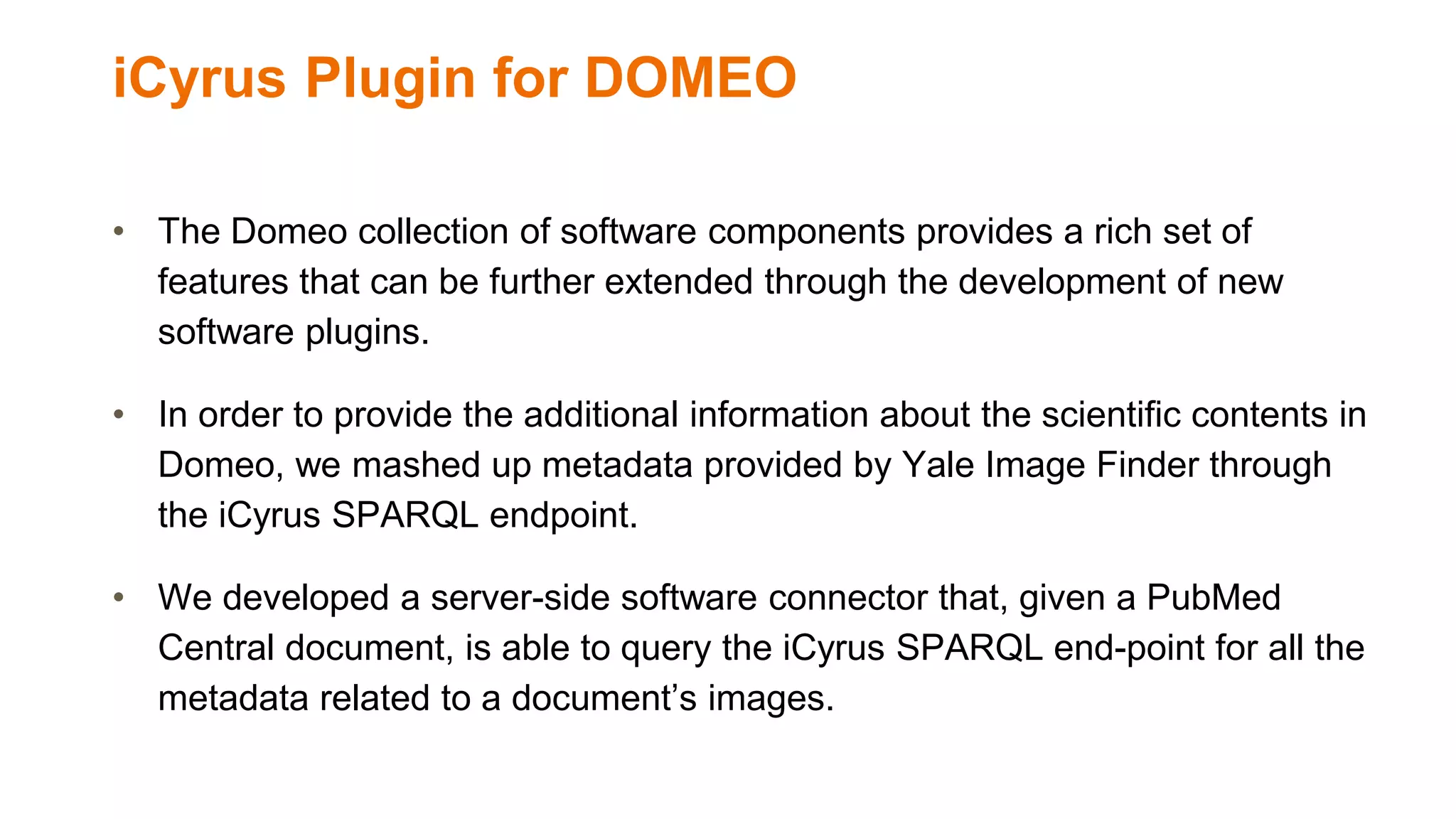
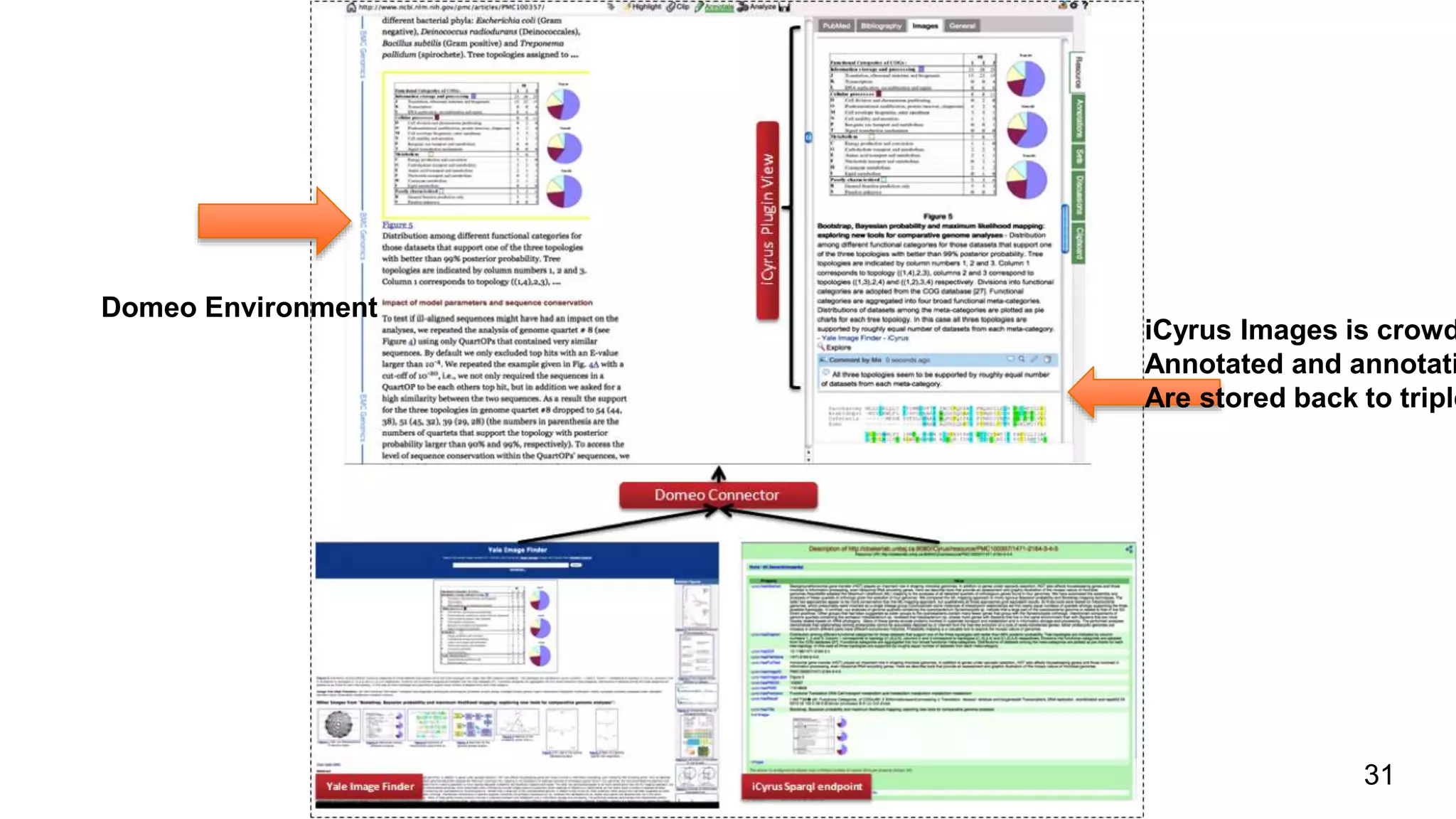
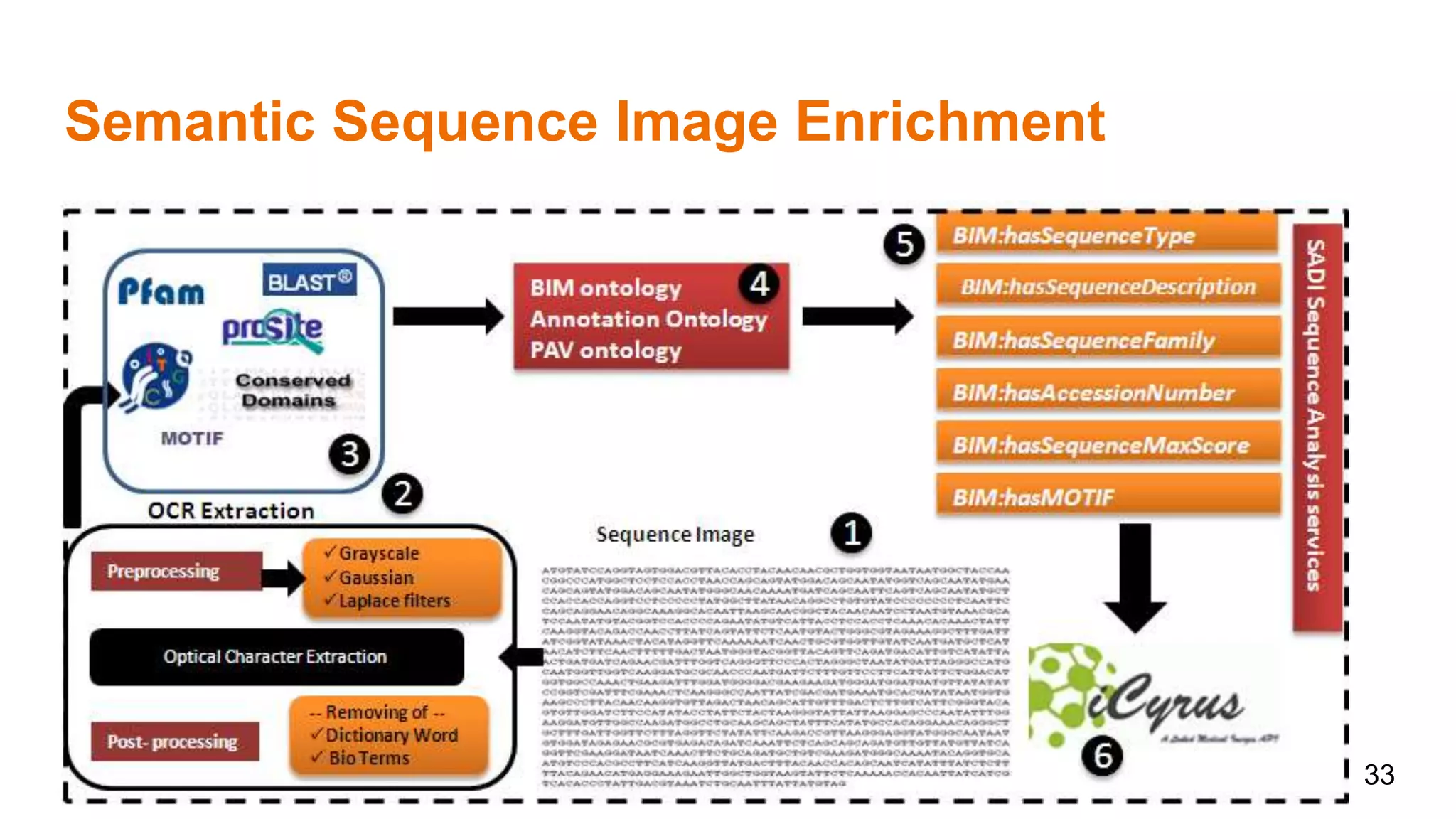
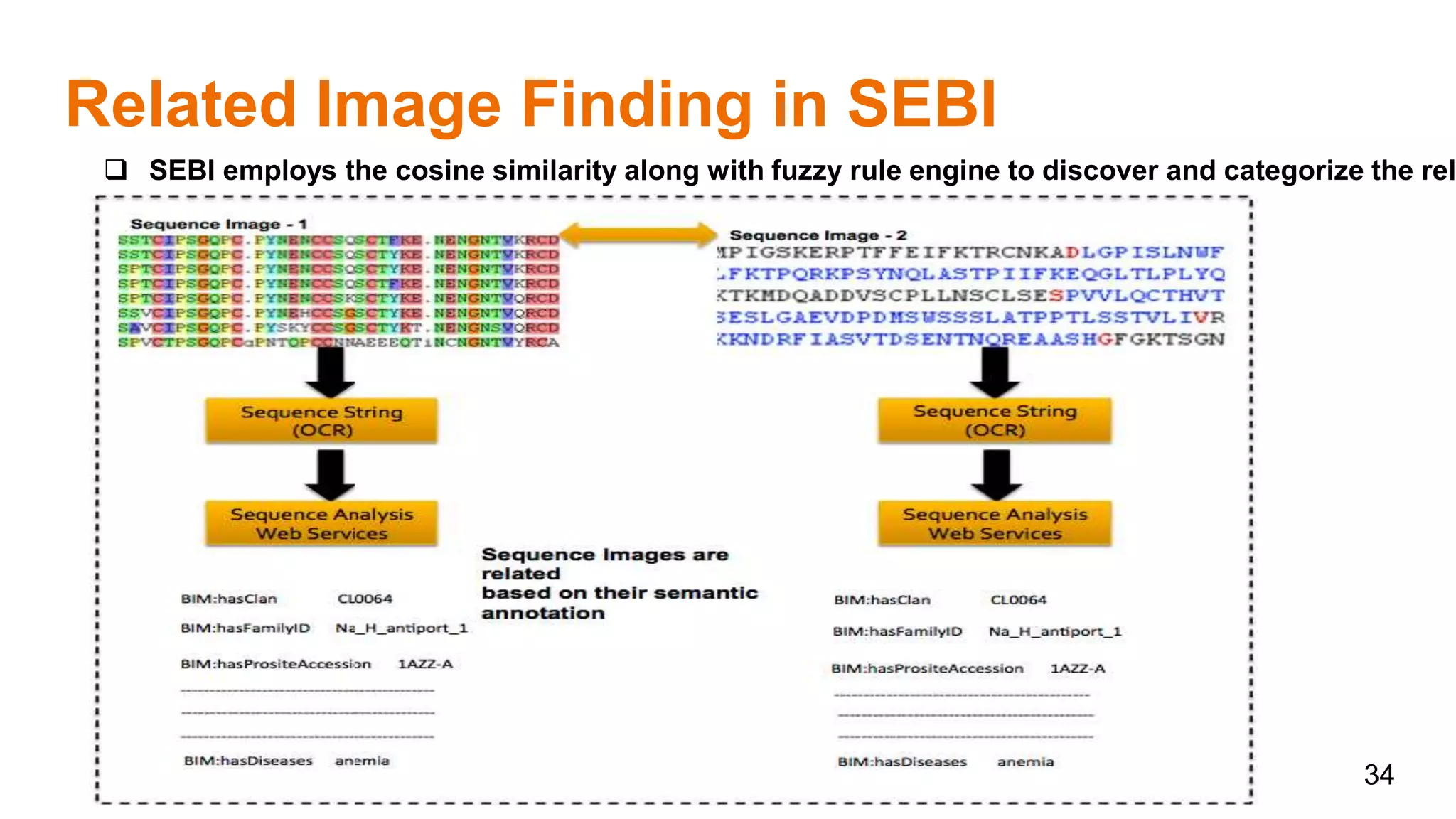
The document discusses the development of a semantic framework, called icyrus, for efficiently discovering and integrating biomedical images, addressing current challenges related to data management and interoperability in biomedical research. It emphasizes the importance of semantic APIs for enhancing access to biomedical images and proposes solutions to improve knowledge extraction and reuse through linked data services. The framework aims to support clinicians, researchers, educators, and patients in their search for relevant biomedical imagery and information.