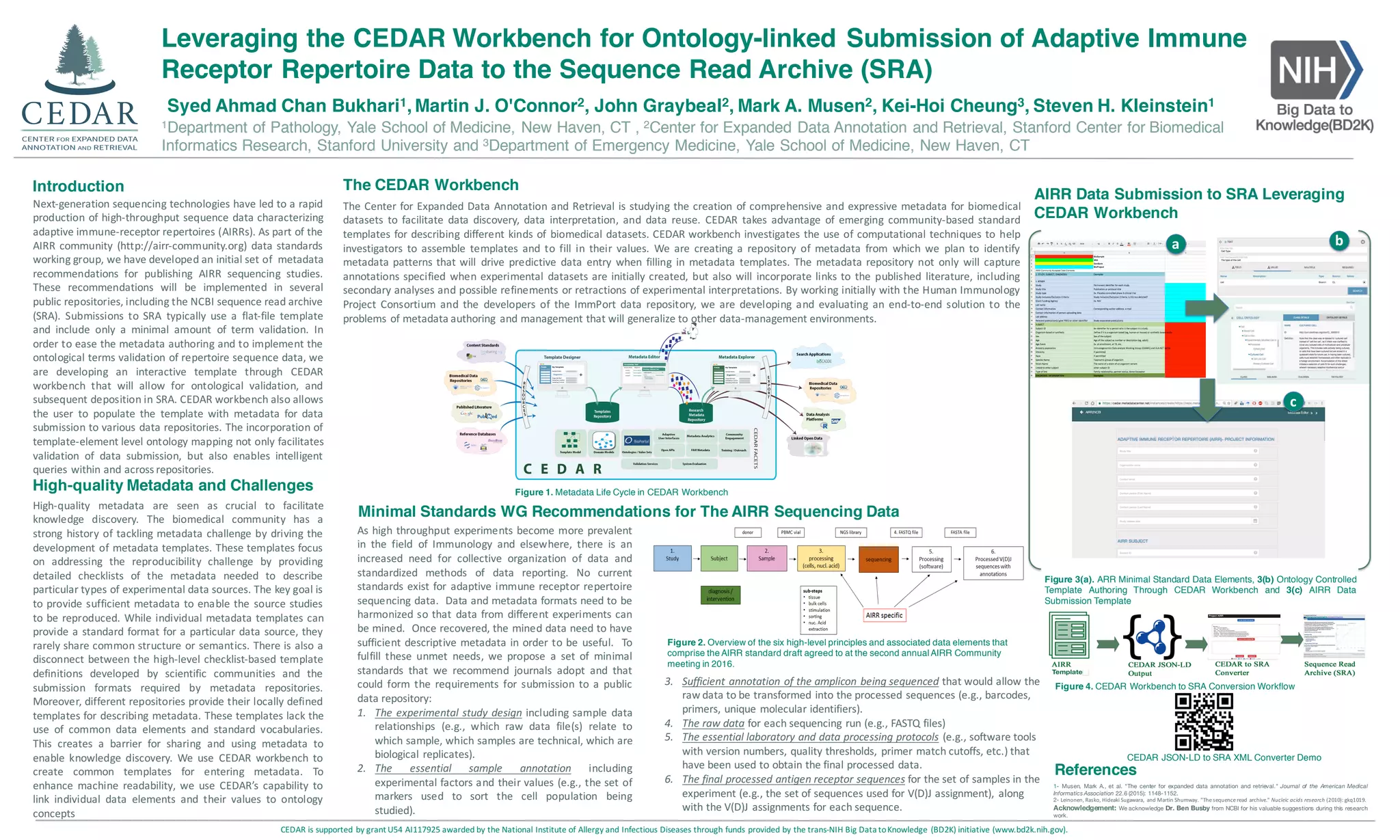
This document discusses the development of metadata standards and templates for publishing adaptive immune receptor repertoire (AIRR) sequencing data. It describes the CEDAR workbench, which allows users to create metadata templates using controlled vocabularies and ontologies. Templates created in CEDAR can then be used to submit AIRR data to repositories like the Sequence Read Archive in a standardized, machine-readable way. This facilitates knowledge discovery by enabling intelligent queries within and across repositories.