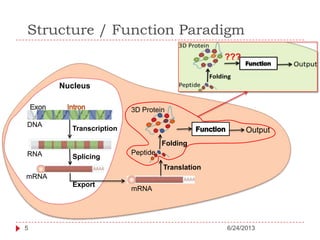
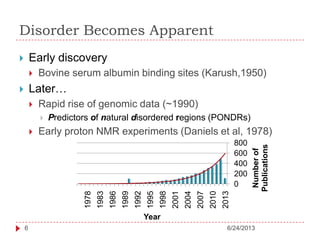
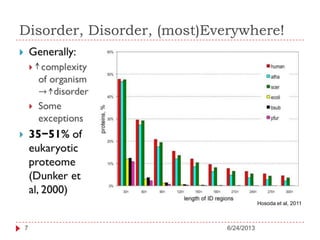
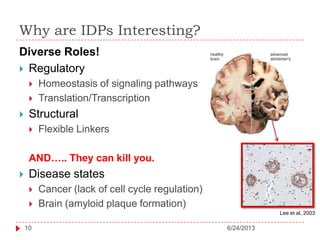
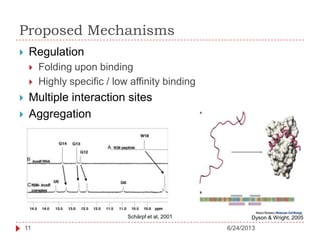
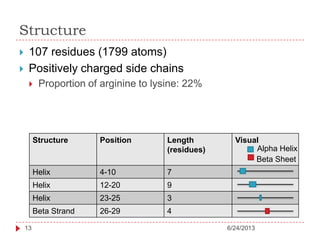
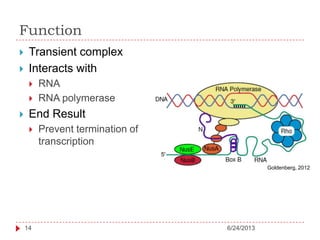
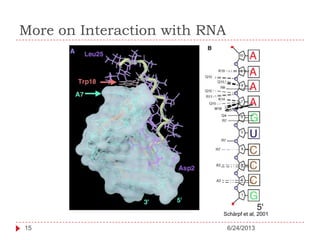
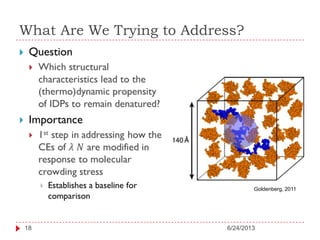
The document discusses intrinsically disordered proteins (IDPs), highlighting their structural characteristics and significant roles in biological processes such as regulation and disease. It notes a historical underestimation of protein disorder due to experimental biases and advancements in genomic data since the 1990s that helped in discovering IDPs. The document also outlines the implications of IDPs in various diseases, including cancer and Alzheimer’s, and aims to provide insights into their atomistic properties.