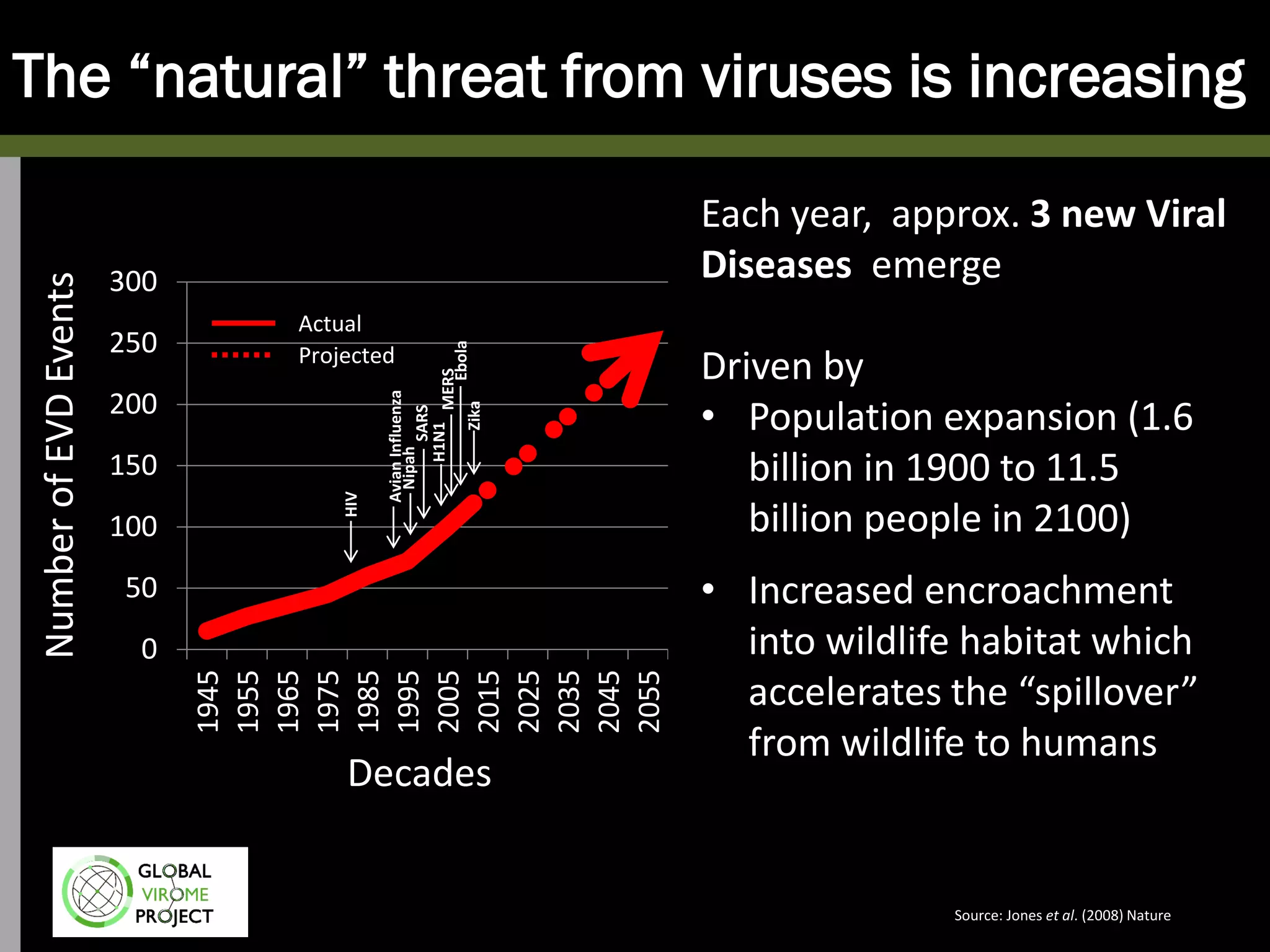
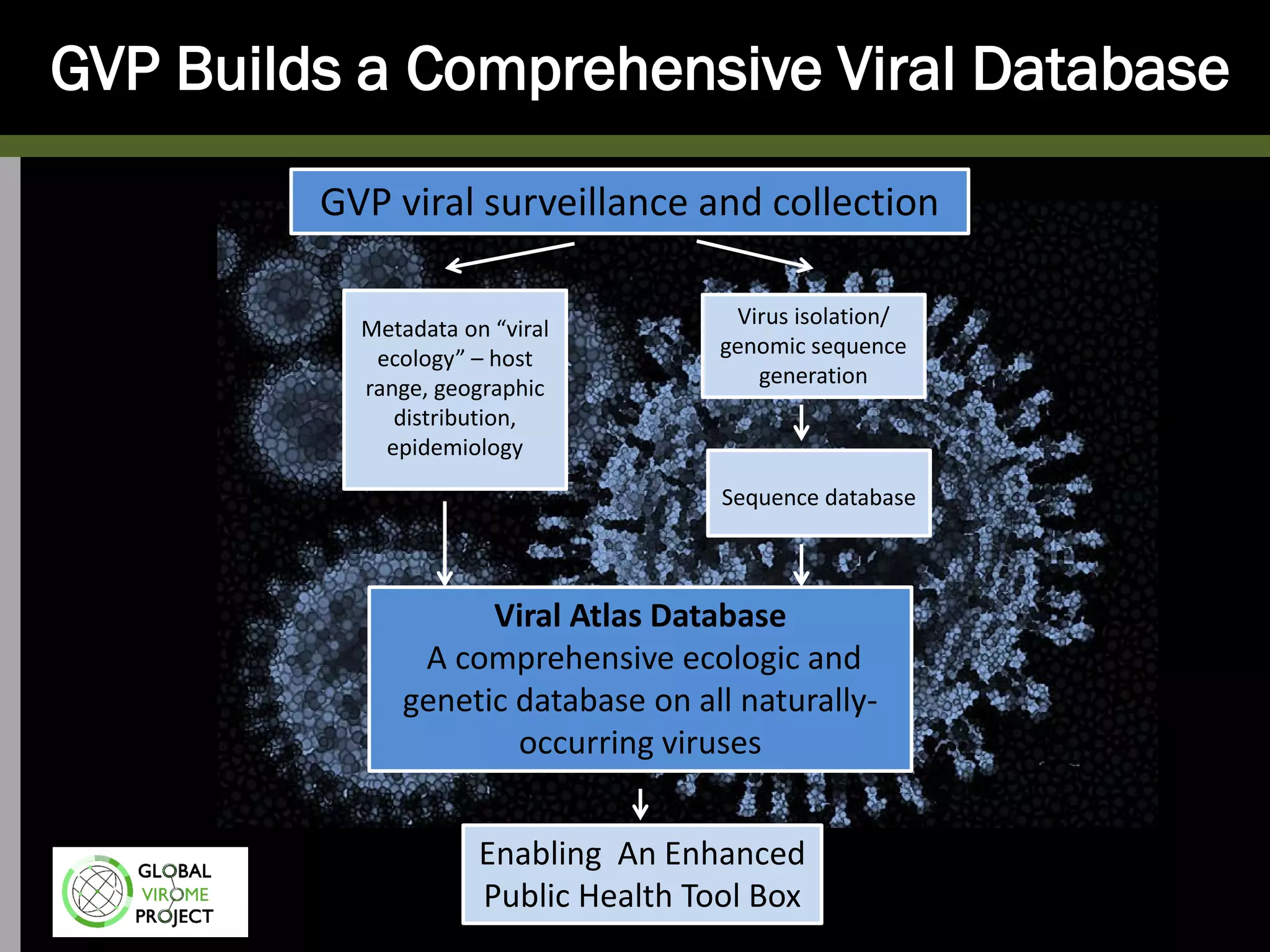
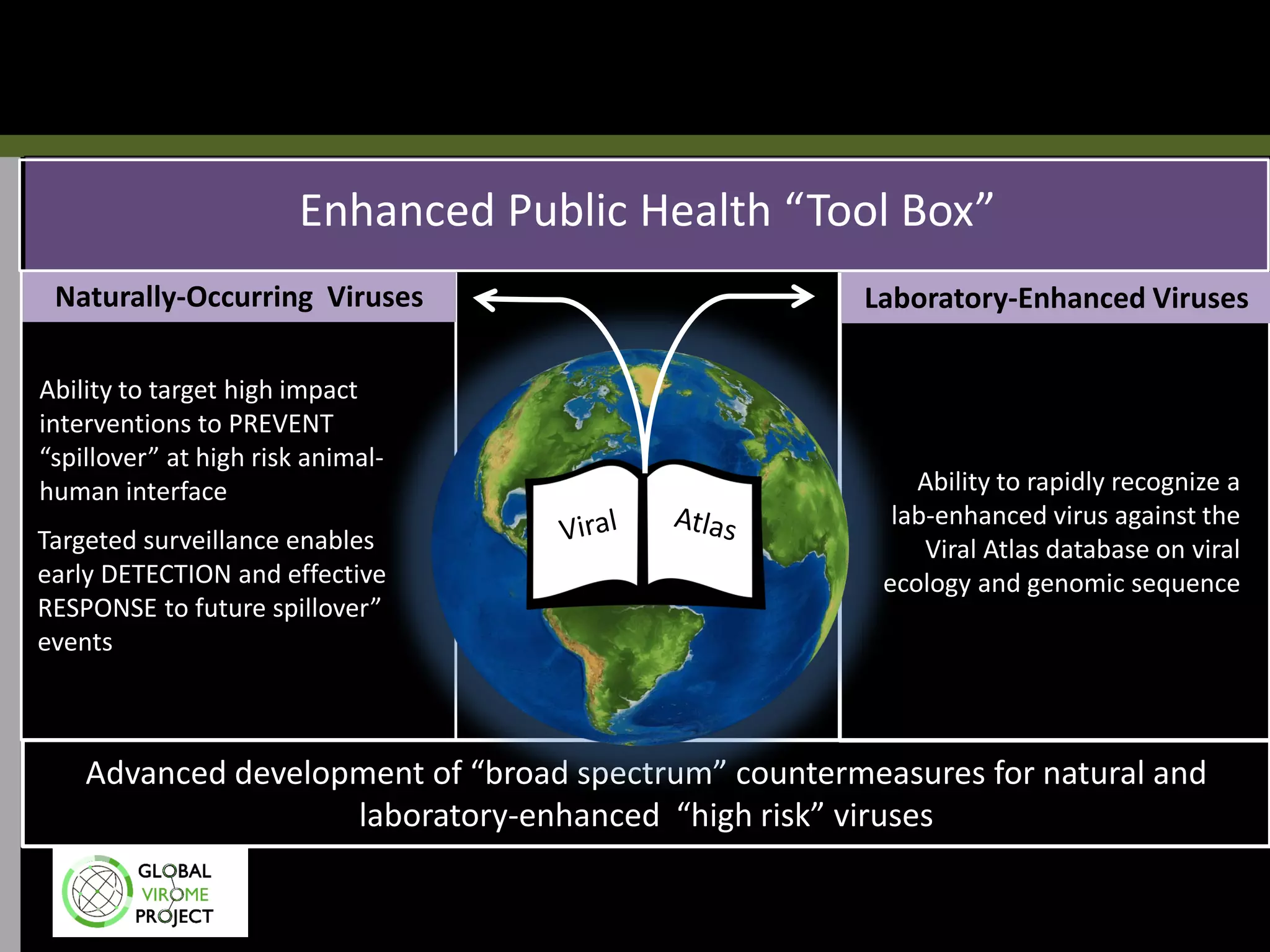
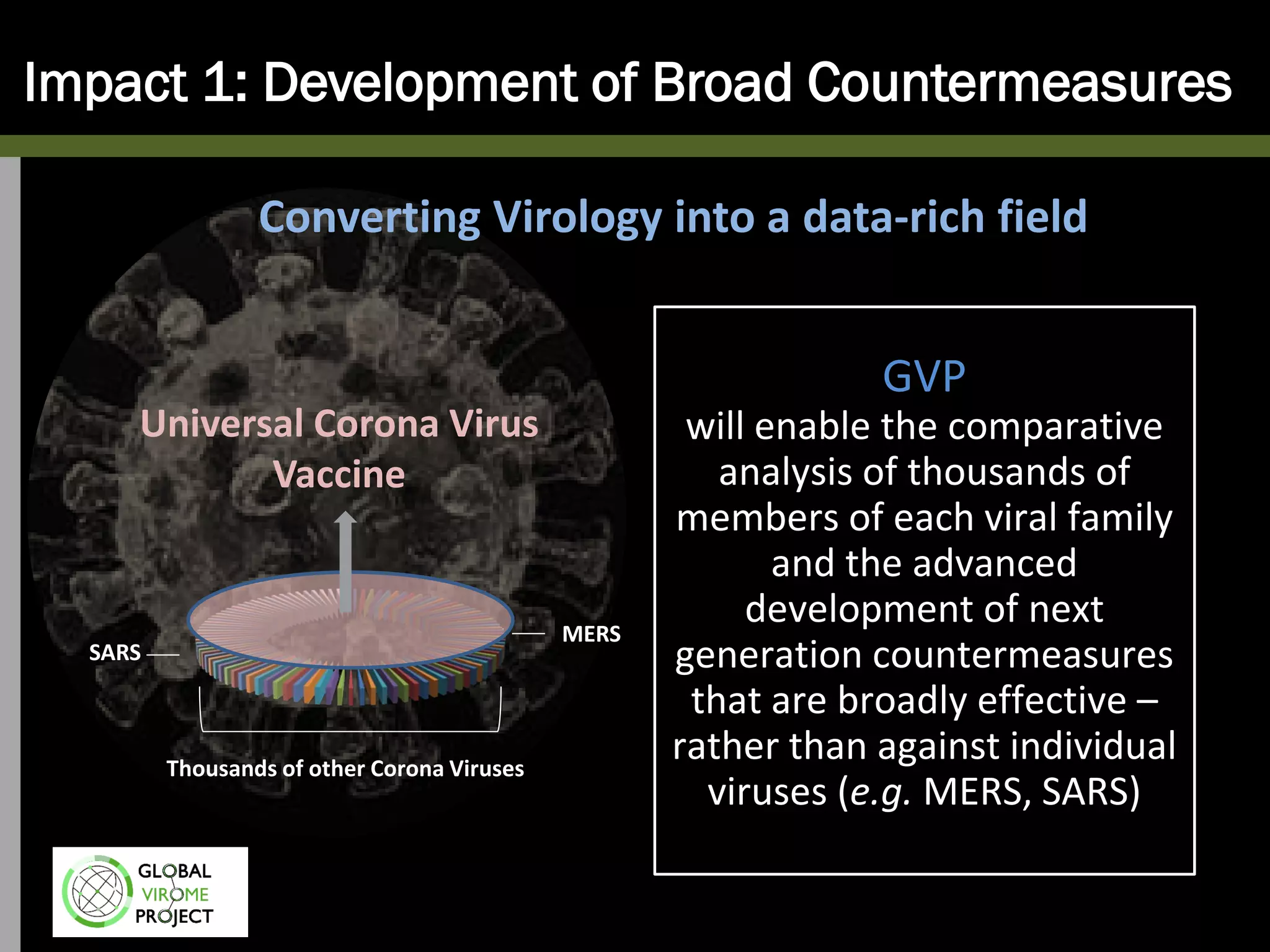
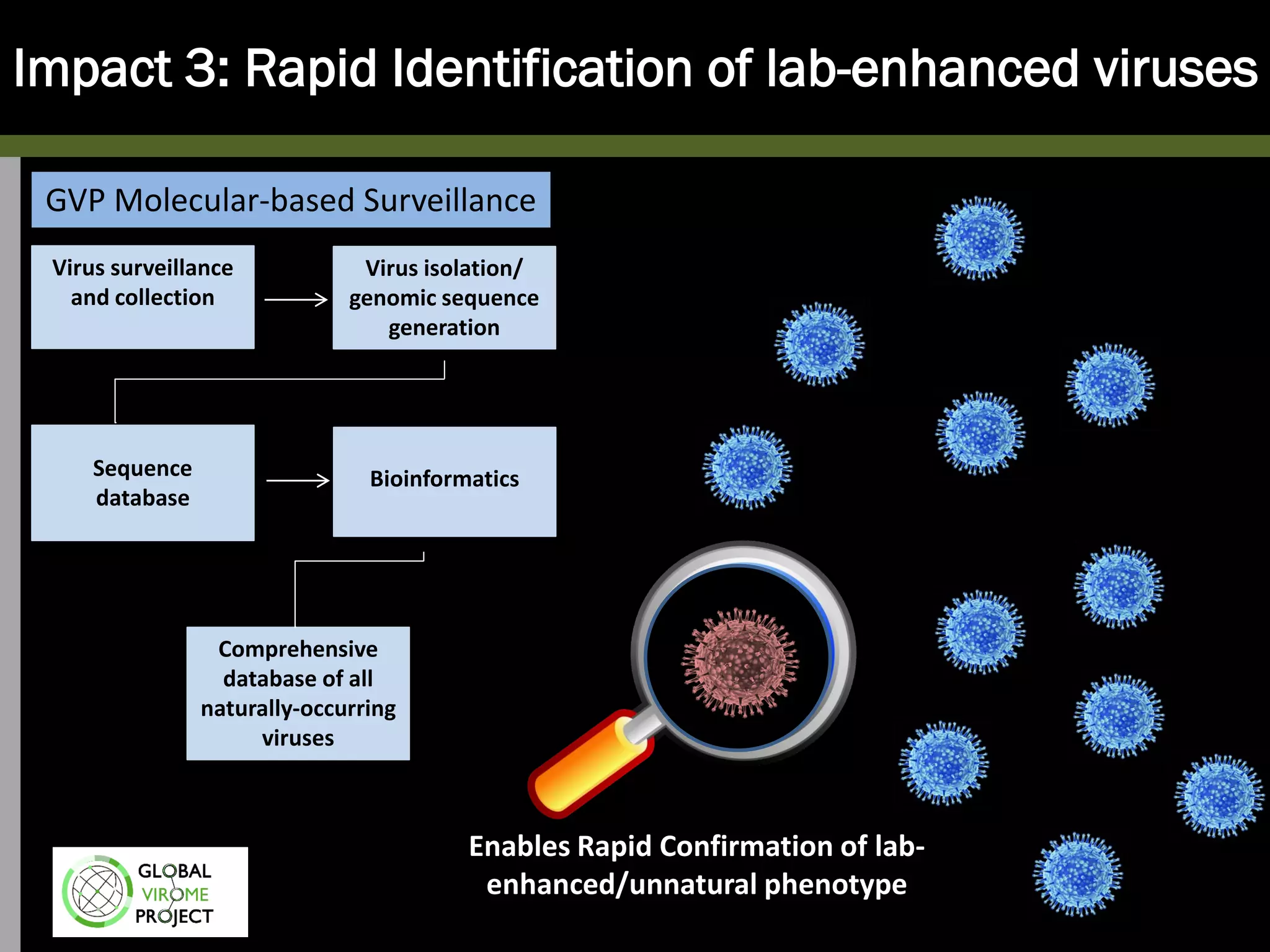
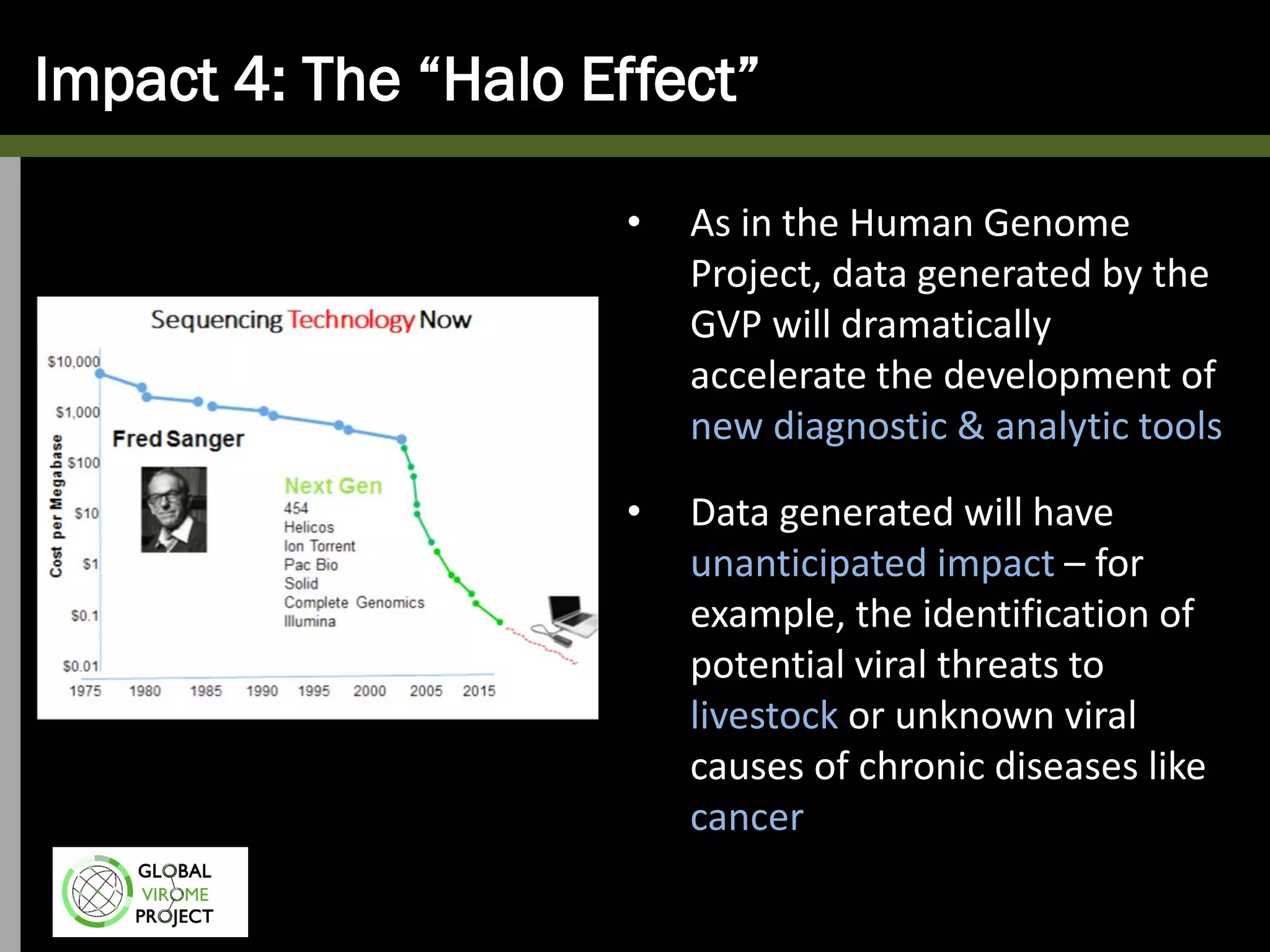
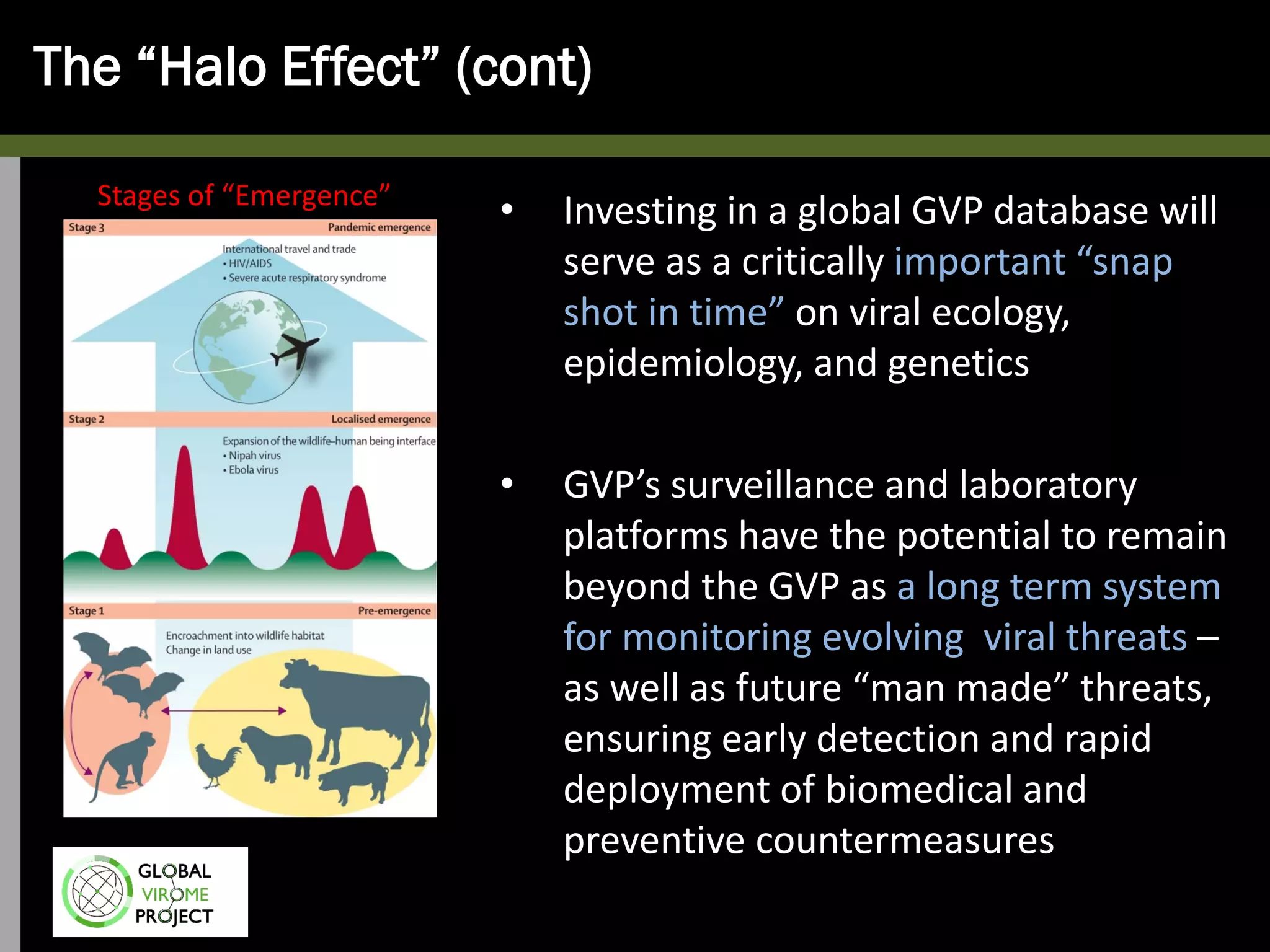
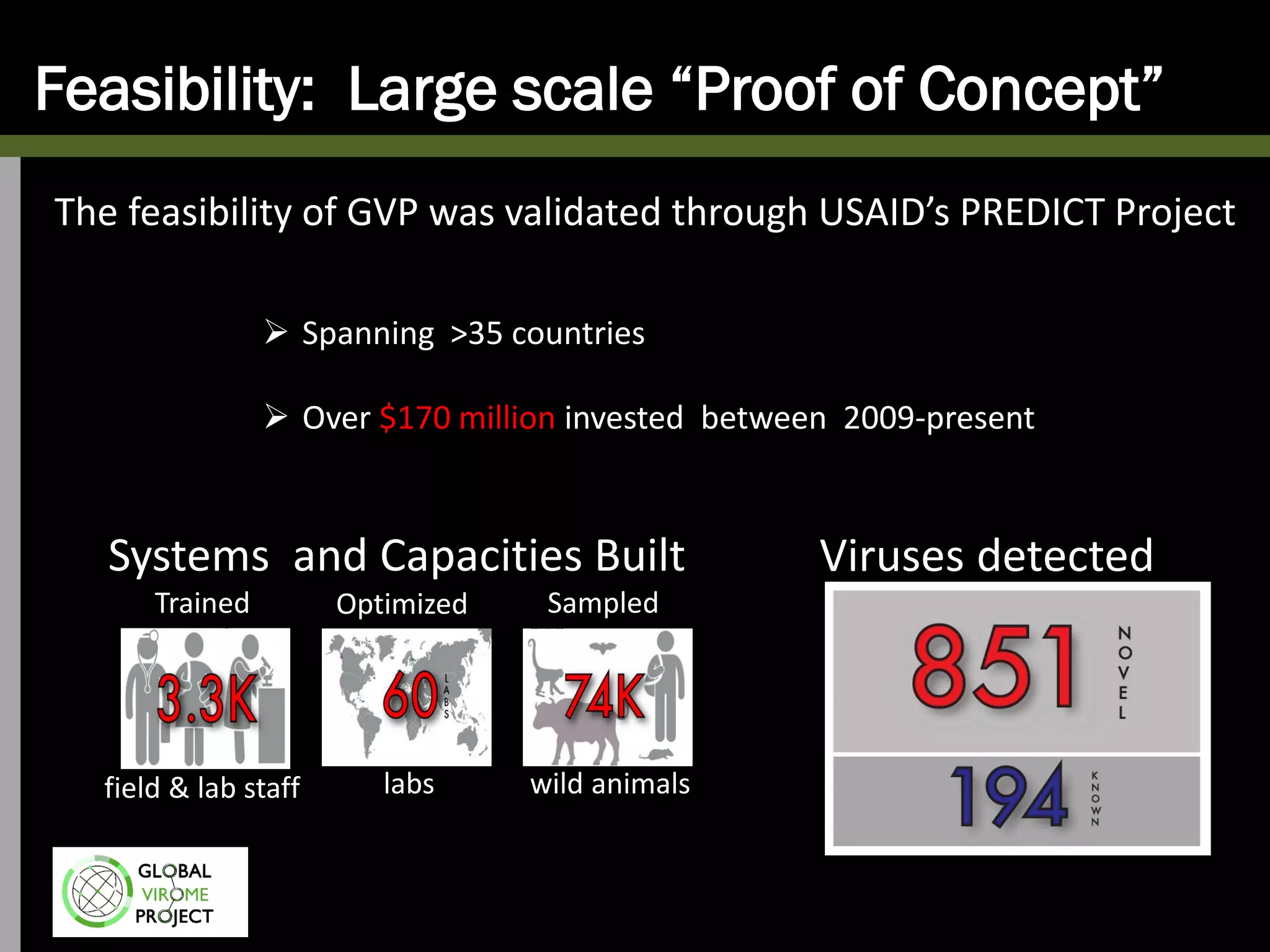
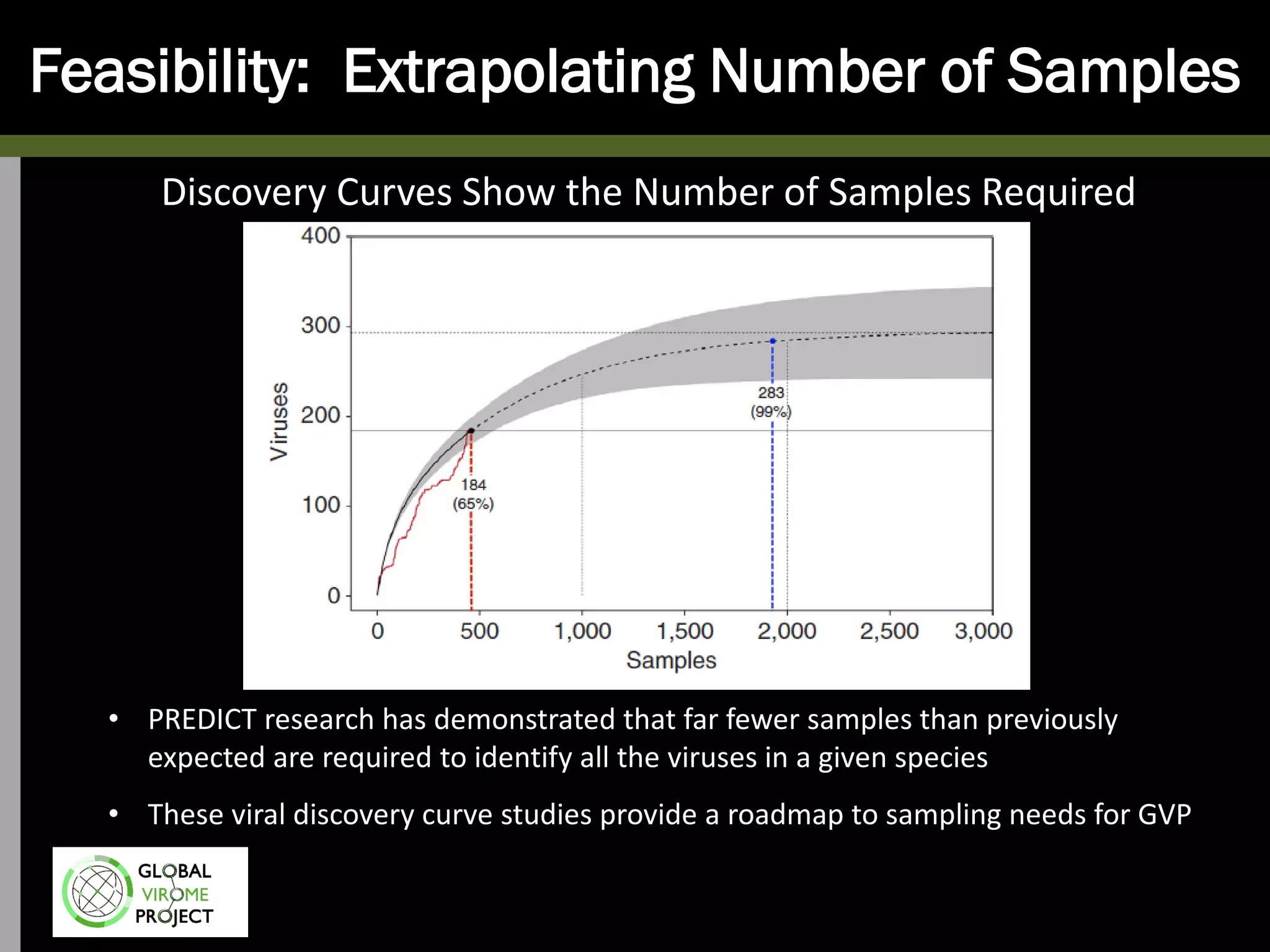
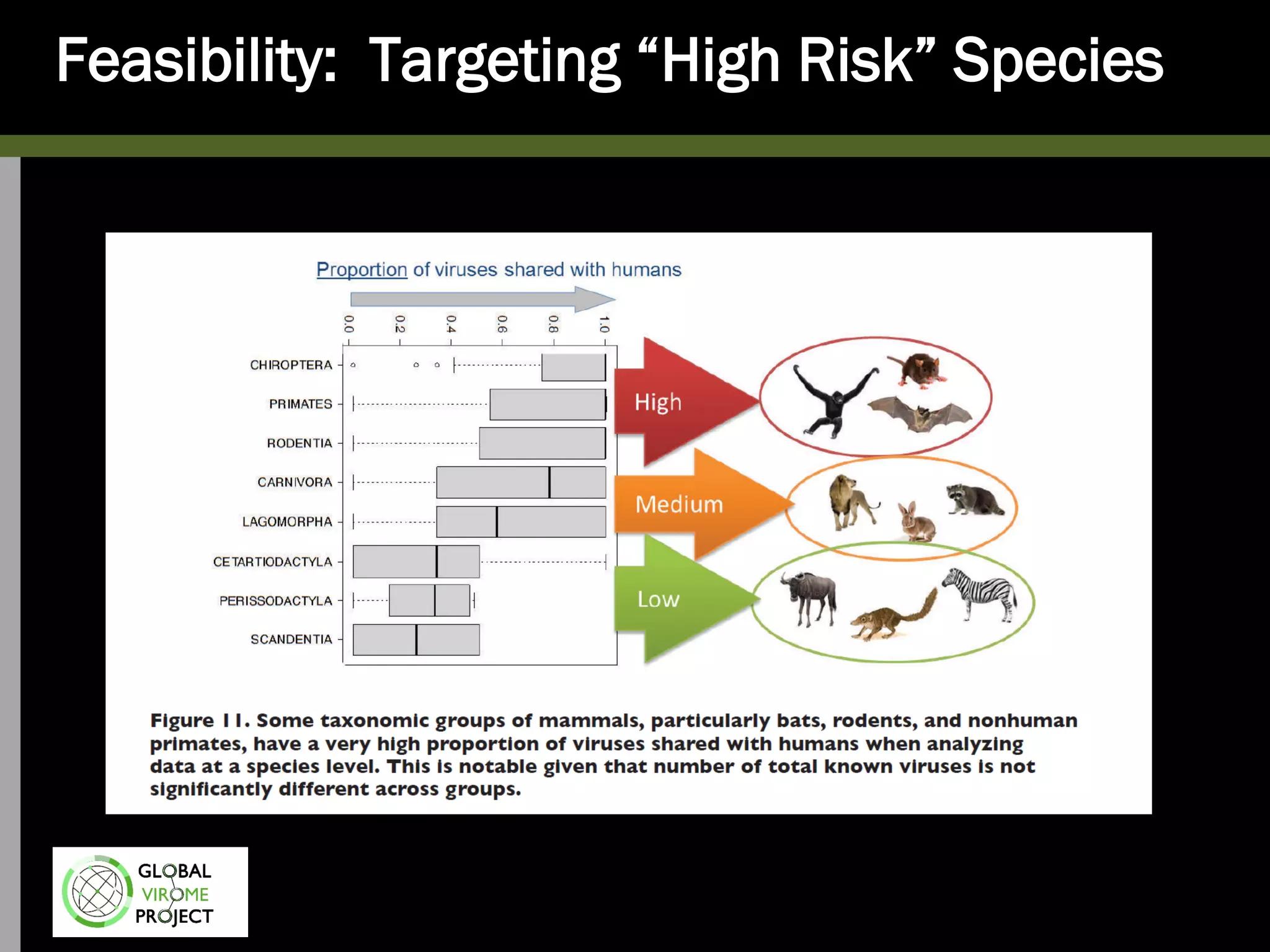
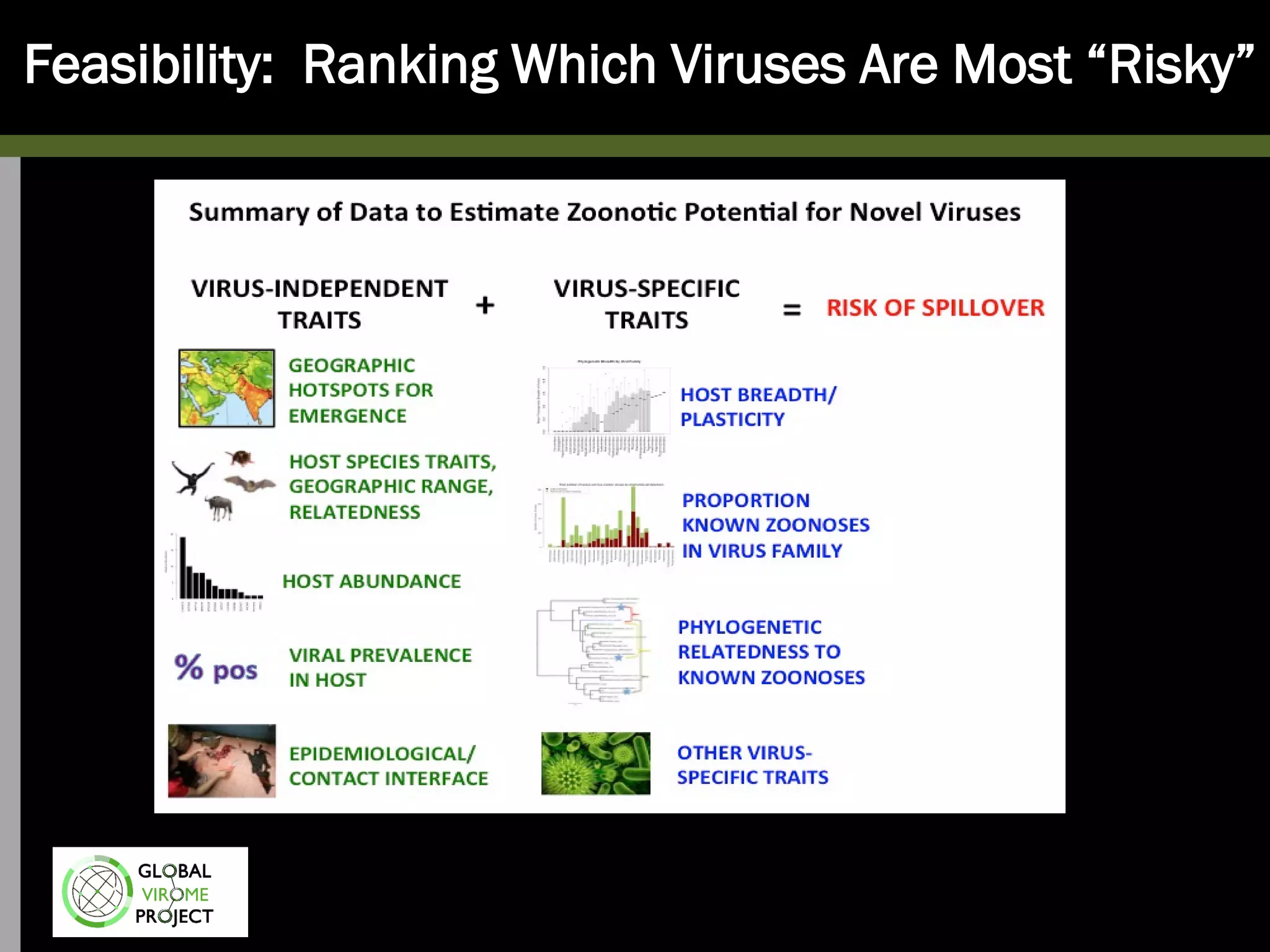
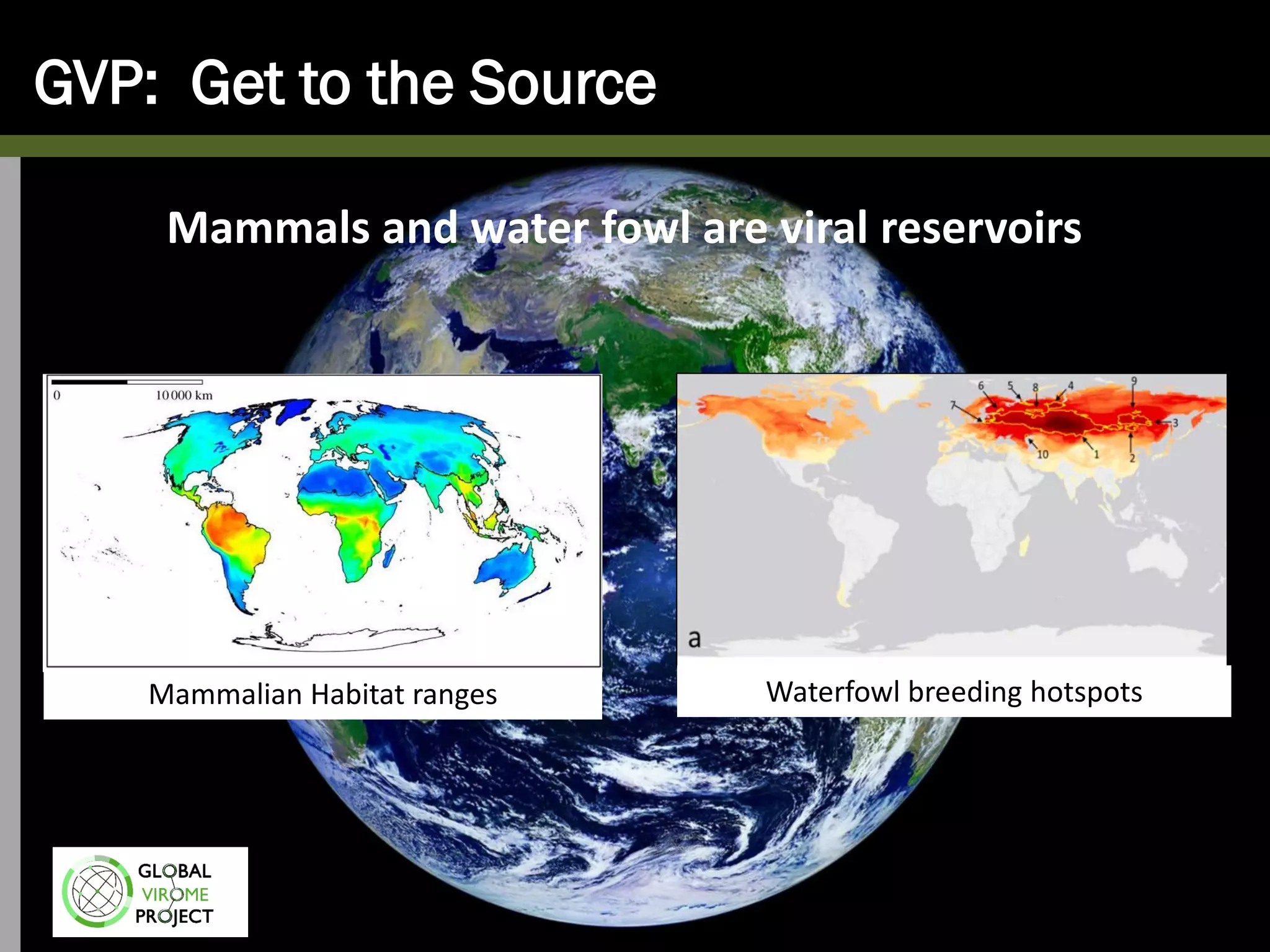
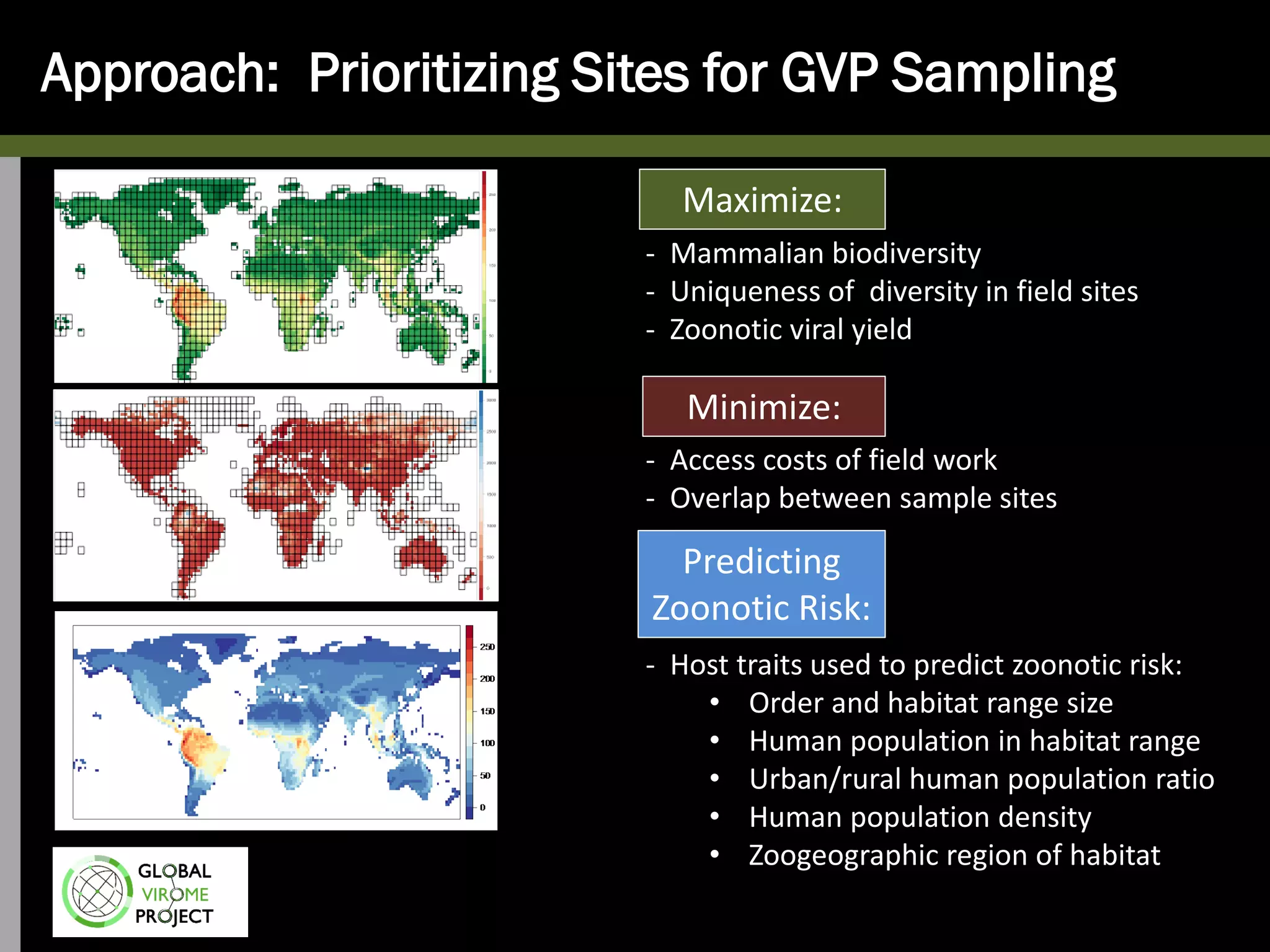
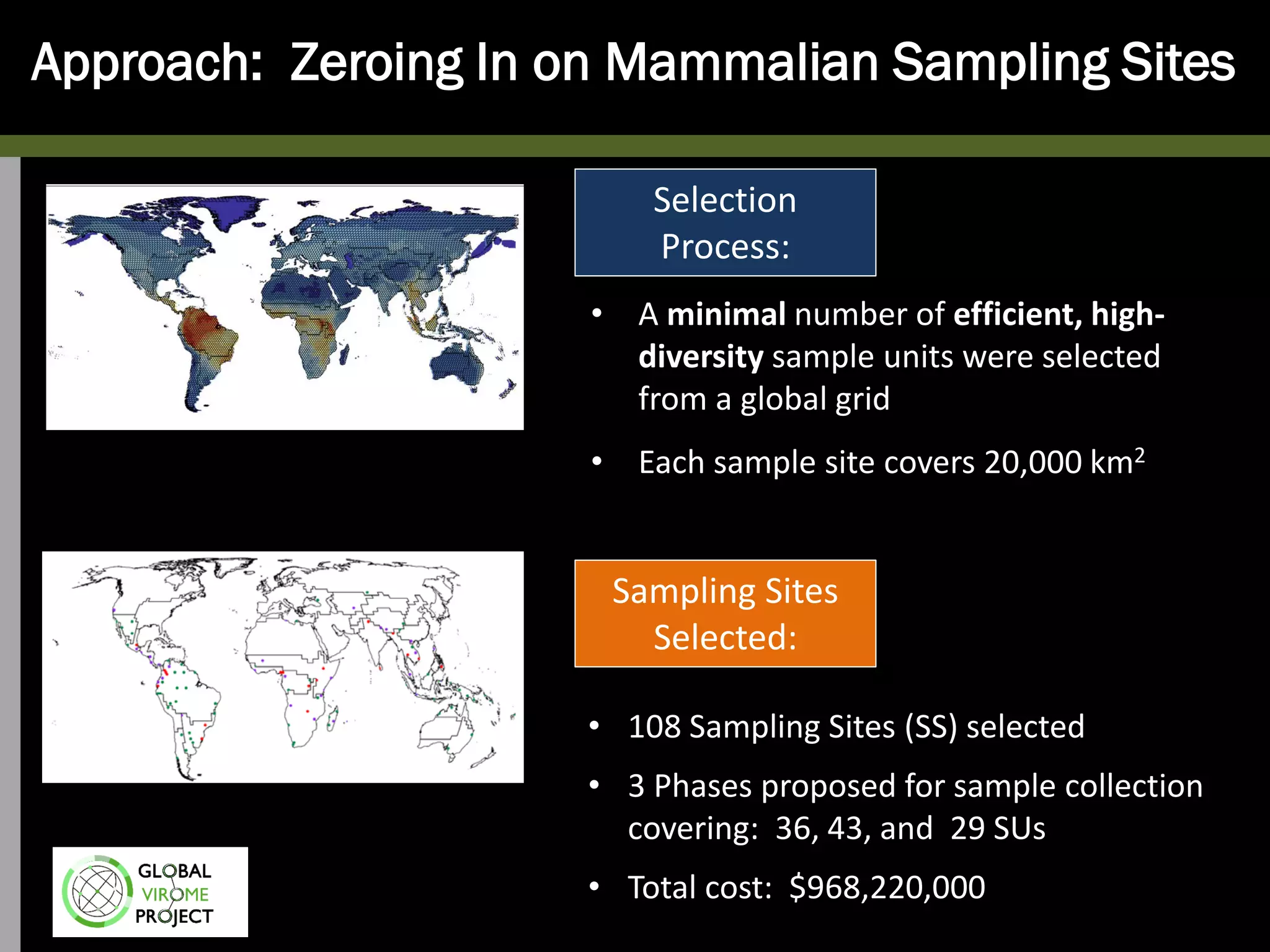
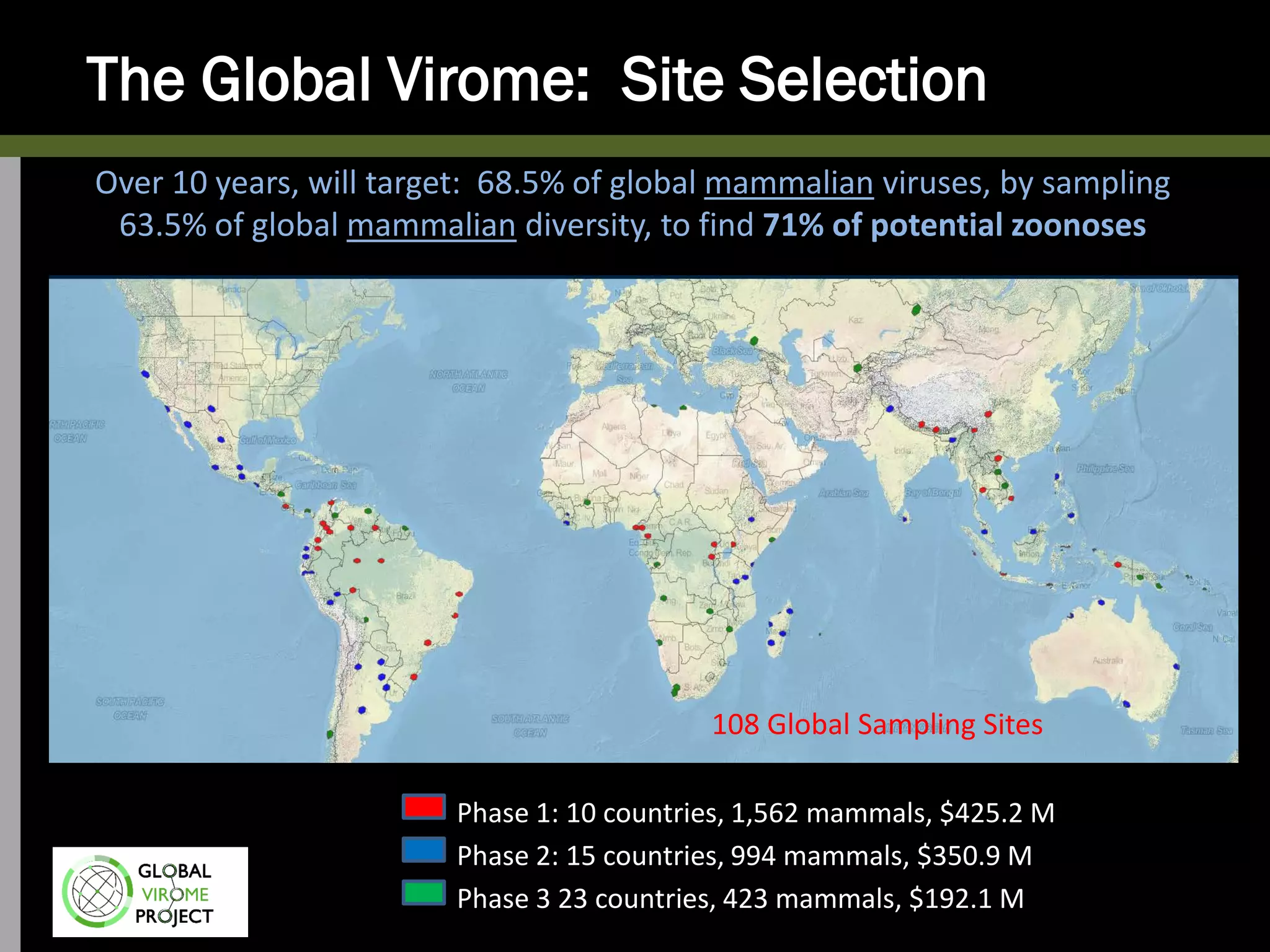
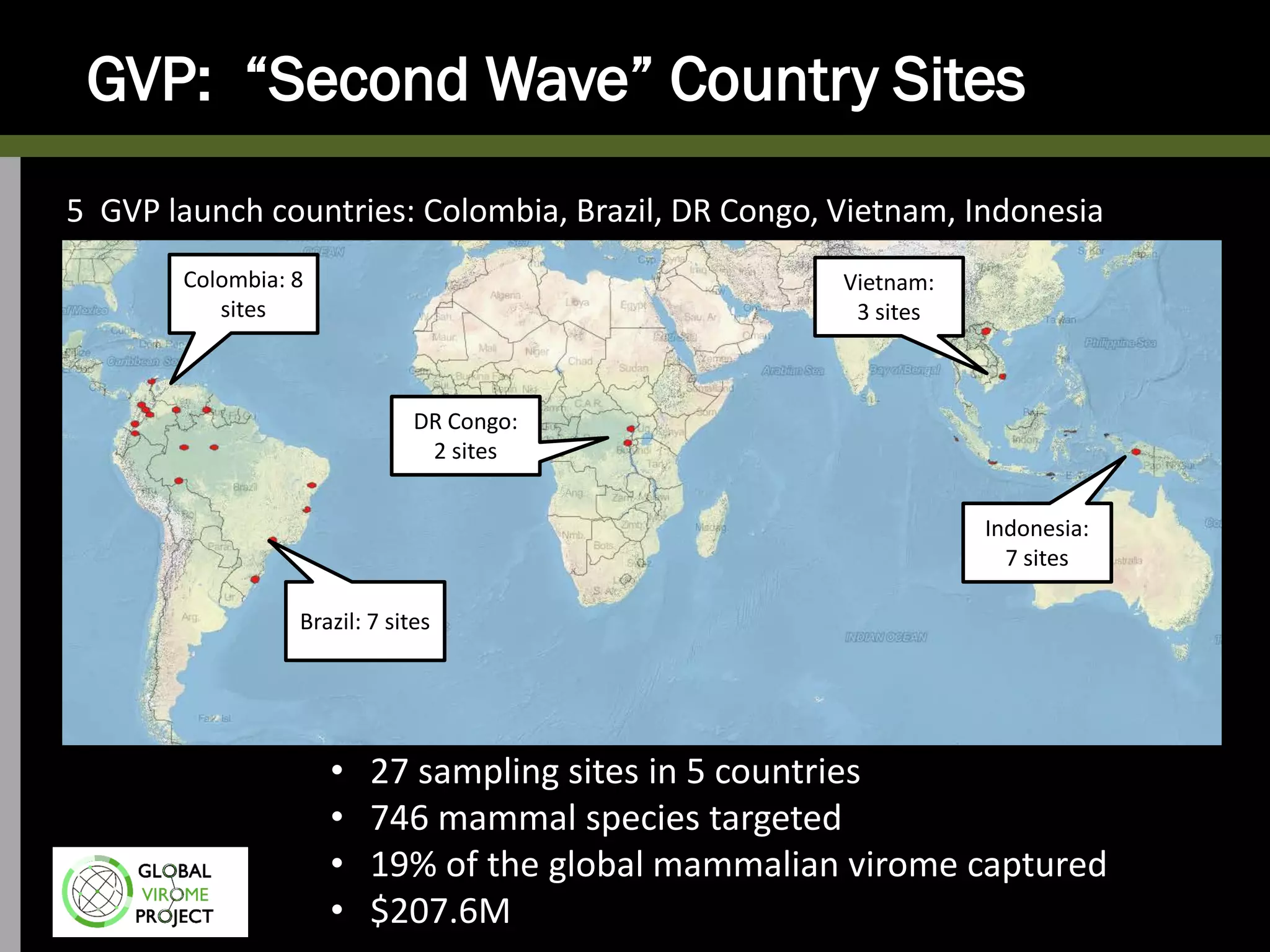
The Global Virome Project is a 10-year global effort to identify and characterize naturally occurring viruses with pandemic potential. It aims to build a comprehensive database of the estimated 1.6 million viral species circulating in mammals and waterfowl. This will allow researchers to develop broad-spectrum countermeasures against future zoonotic viruses and identify high-risk viruses to prevent spillover. The project will sample viruses in 108 sites across 63 countries over 10 years, prioritizing countries and species based on viral discovery rates and zoonotic risk prediction models. The goal is to capture over 85% of the global mammalian virome to transform virology and pandemic preparedness.