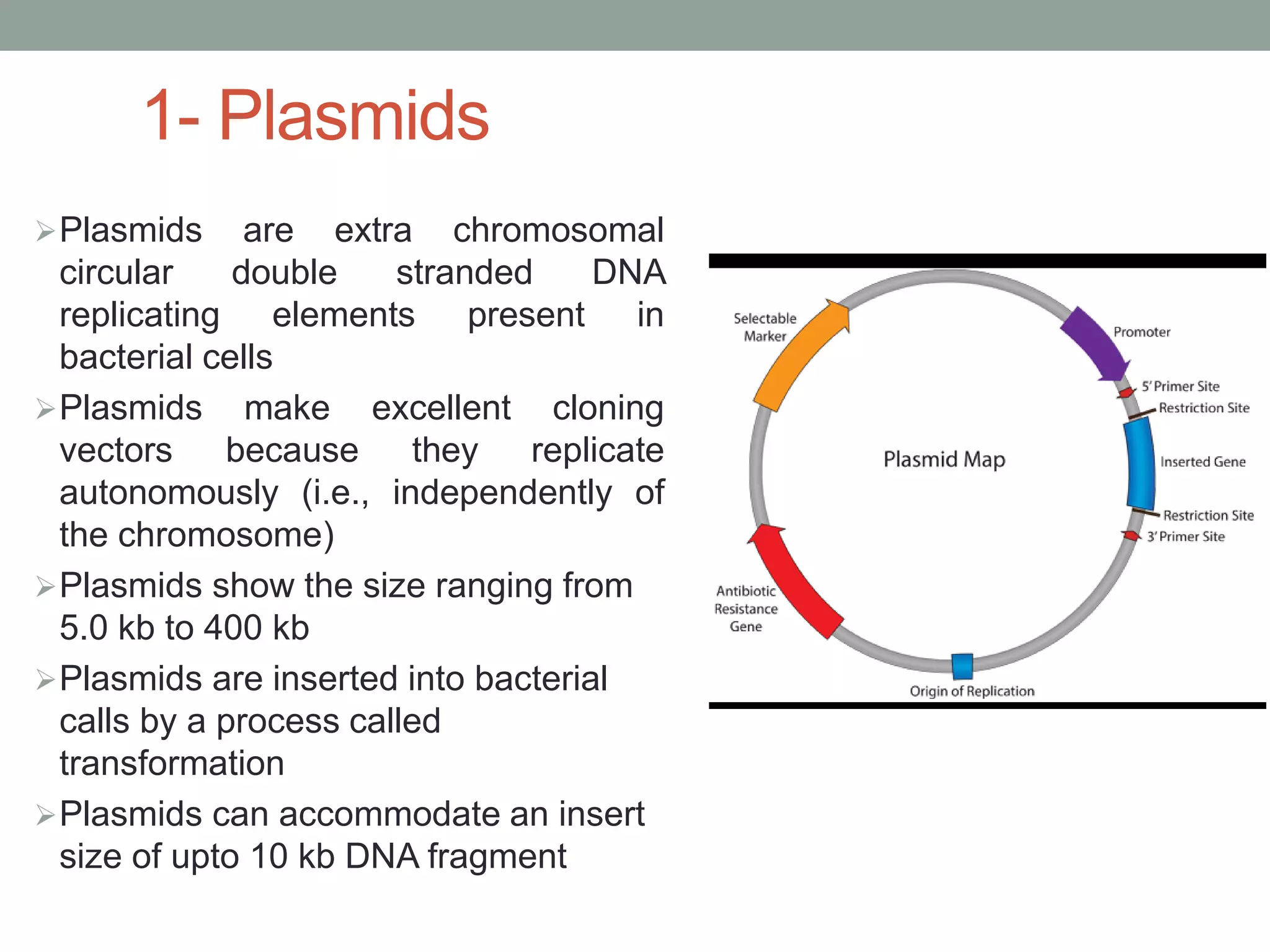
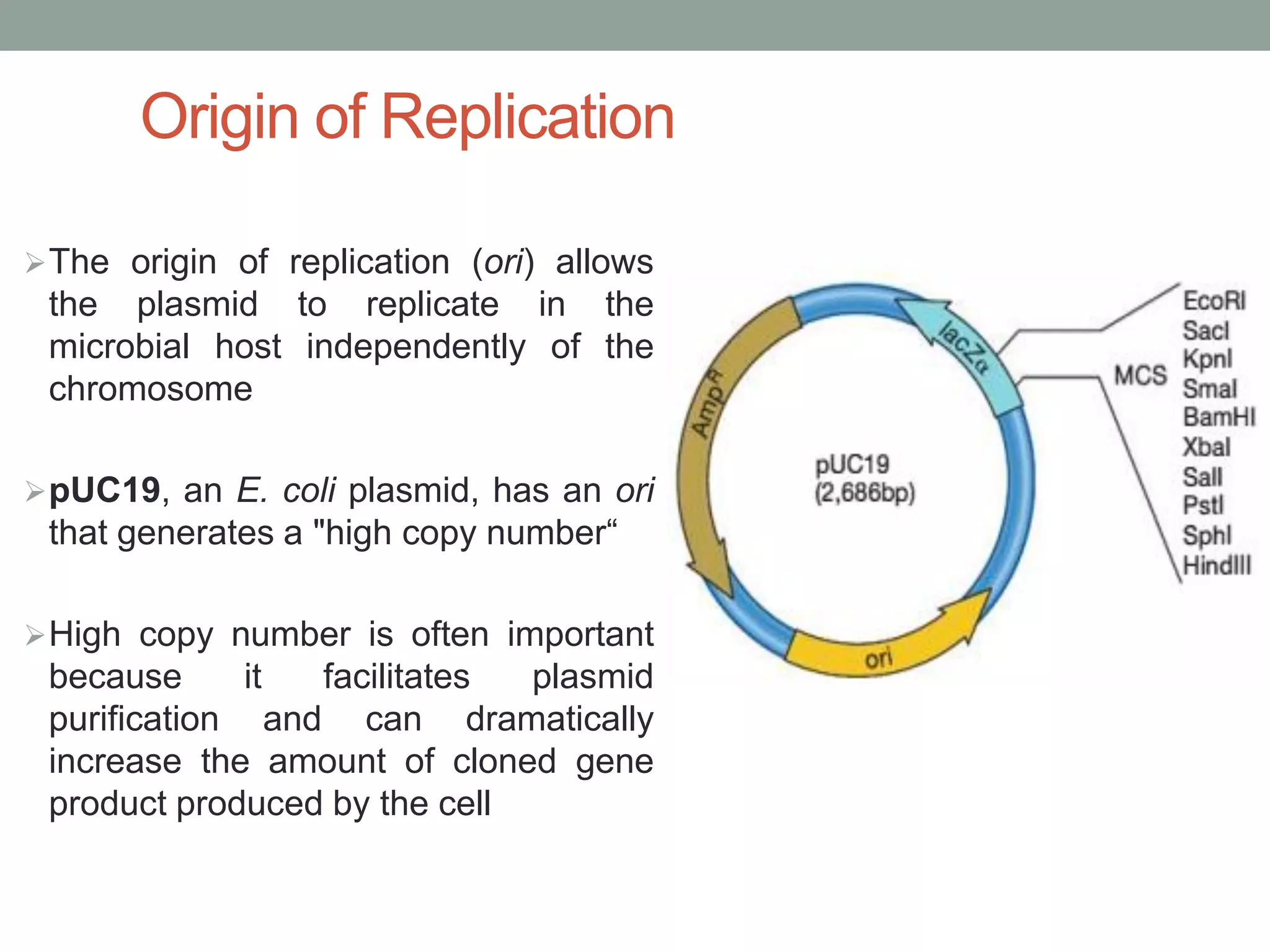
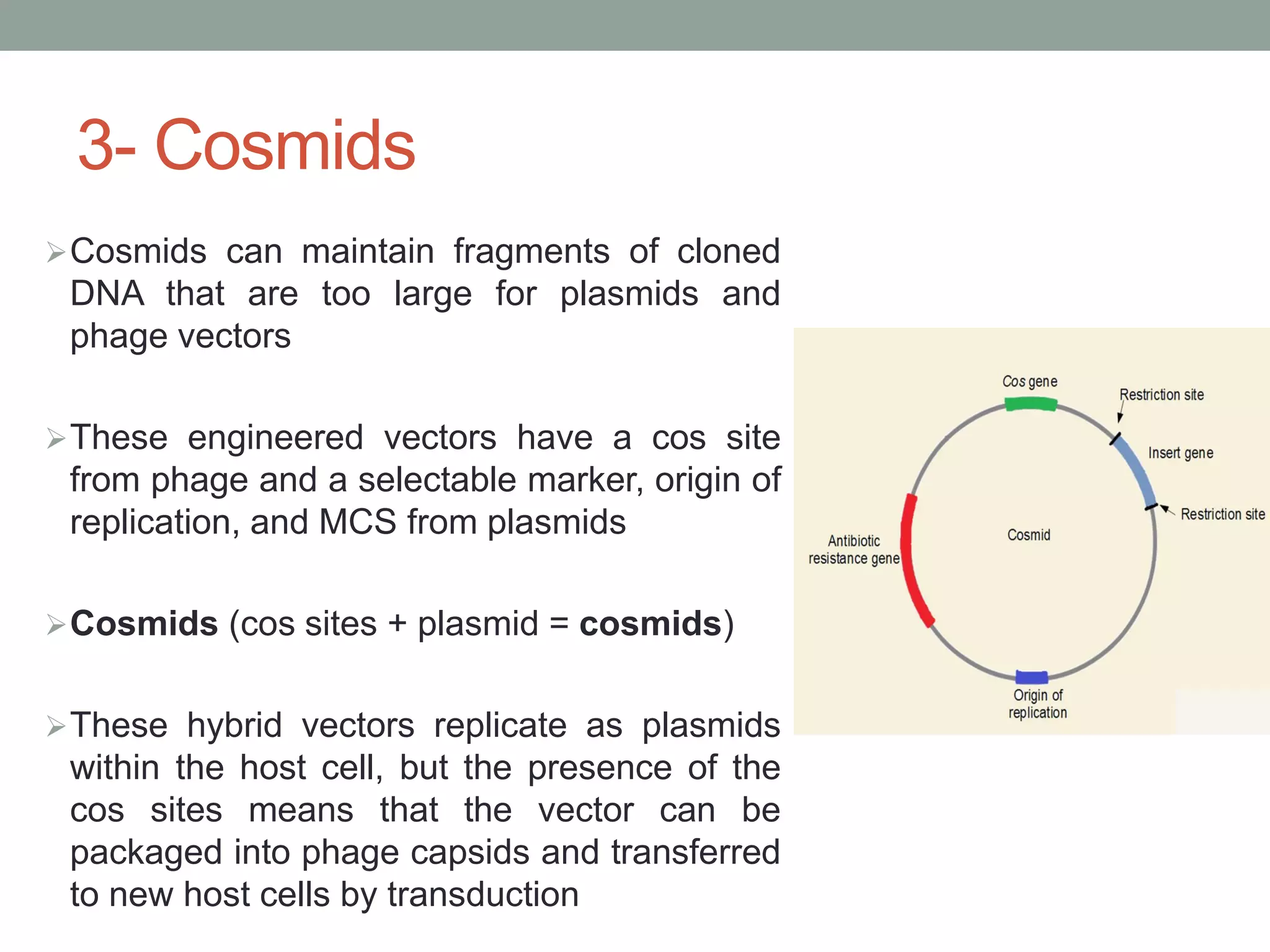
Cloning vectors are DNA molecules used to artificially carry foreign DNA into host cells. The main types are plasmids, phage vectors, cosmids, and artificial chromosomes. Plasmids are extrachromosomal DNA molecules that replicate independently in bacteria. They contain an origin of replication, antibiotic resistance gene, and multicloning site. Phage vectors contain viral genes and can package cloned DNA into viral capsids. Cosmids are hybrid vectors that replicate as plasmids but can also package DNA into phage capsids. Artificial chromosomes like BACs and YACs can clone very large DNA fragments and were important for genome projects.