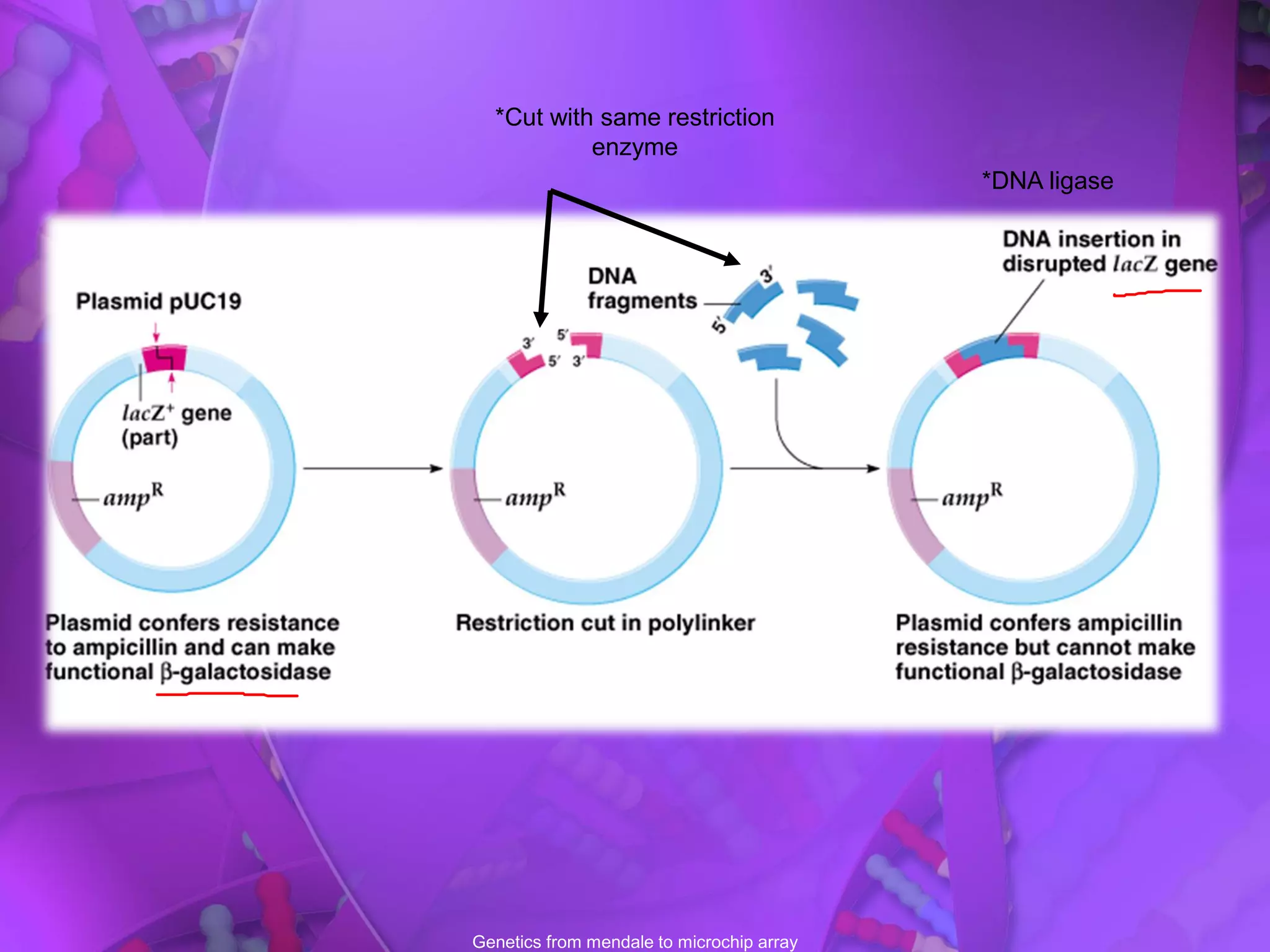
There are six main types of cloning vectors used to insert DNA fragments: plasmid vectors, phage lambda vectors, cosmid vectors, shuttle vectors, yeast artificial chromosomes (YACs), and bacterial artificial chromosomes (BACs). Plasmid vectors are the most commonly used as they can accept DNA fragments up to 10kb and replicate autonomously in bacteria. Lambda phage and cosmid vectors can accept larger fragments up to 52kb, while YACs and BACs are used to clone very large fragments up to 500kb and 200kb respectively.