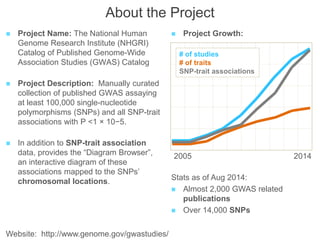
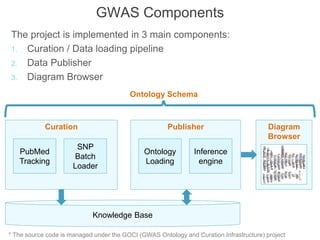
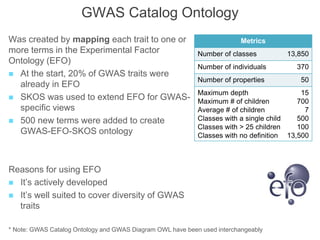
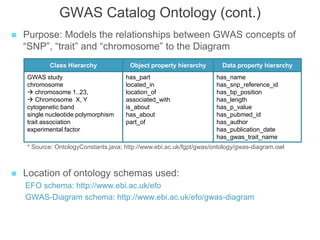
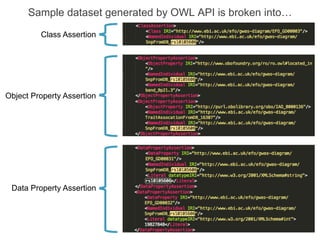
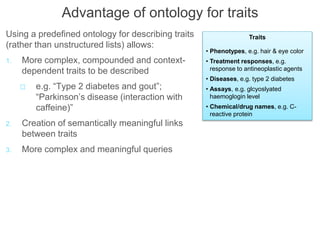
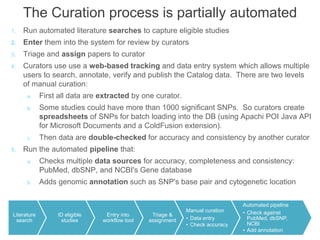
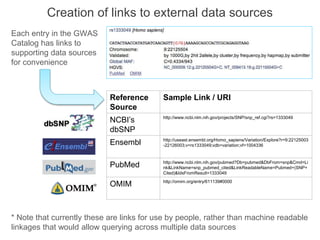
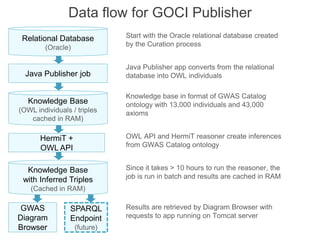
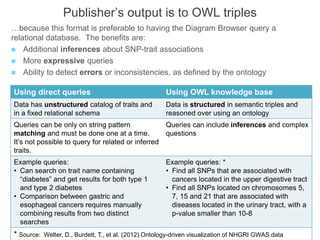
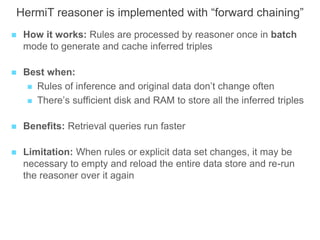
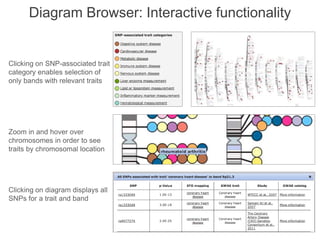
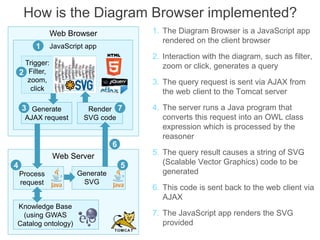
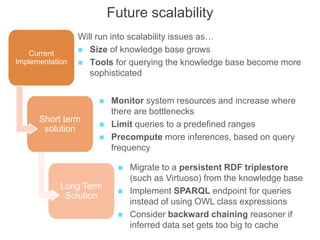
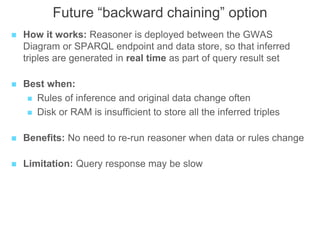
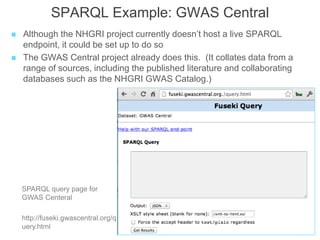
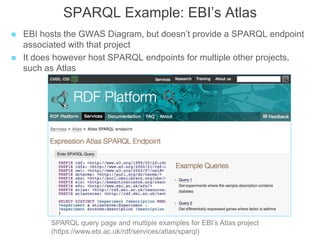
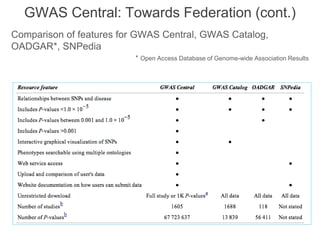
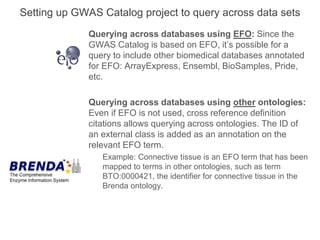
The National Human Genome Research Institute (NHGRI) maintains a catalog of genome-wide association studies (GWAS) that compiles data on single-nucleotide polymorphisms (SNPs) and their associations with traits. The project employs various technologies to facilitate data curation, visualization, and querying, including Java, JavaScript, and the OWL ontology, and features an interactive diagram browser to explore SNP-trait associations. Additionally, future enhancements aim to improve scalability and the querying process through machine learning and natural language processing techniques.