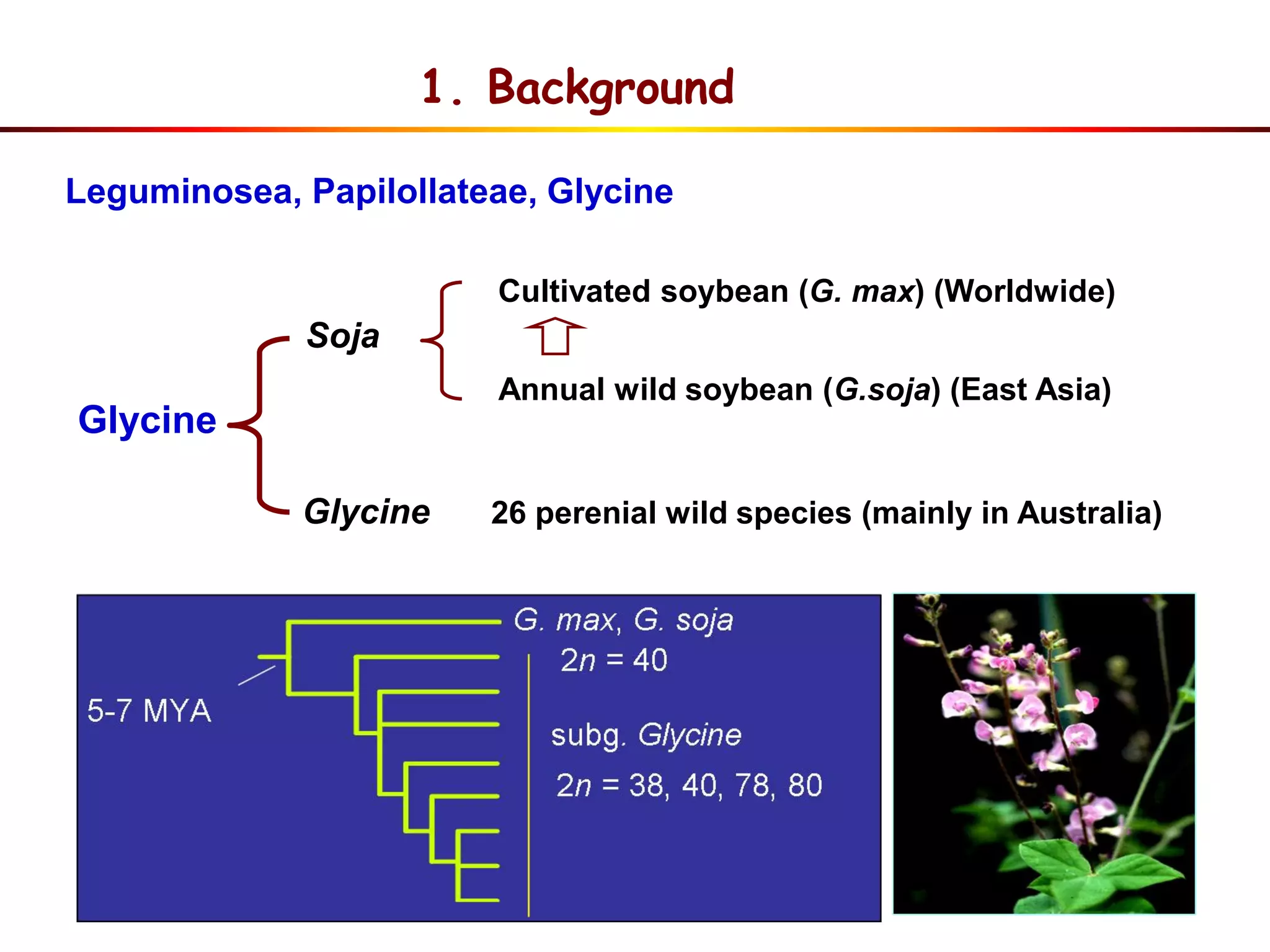
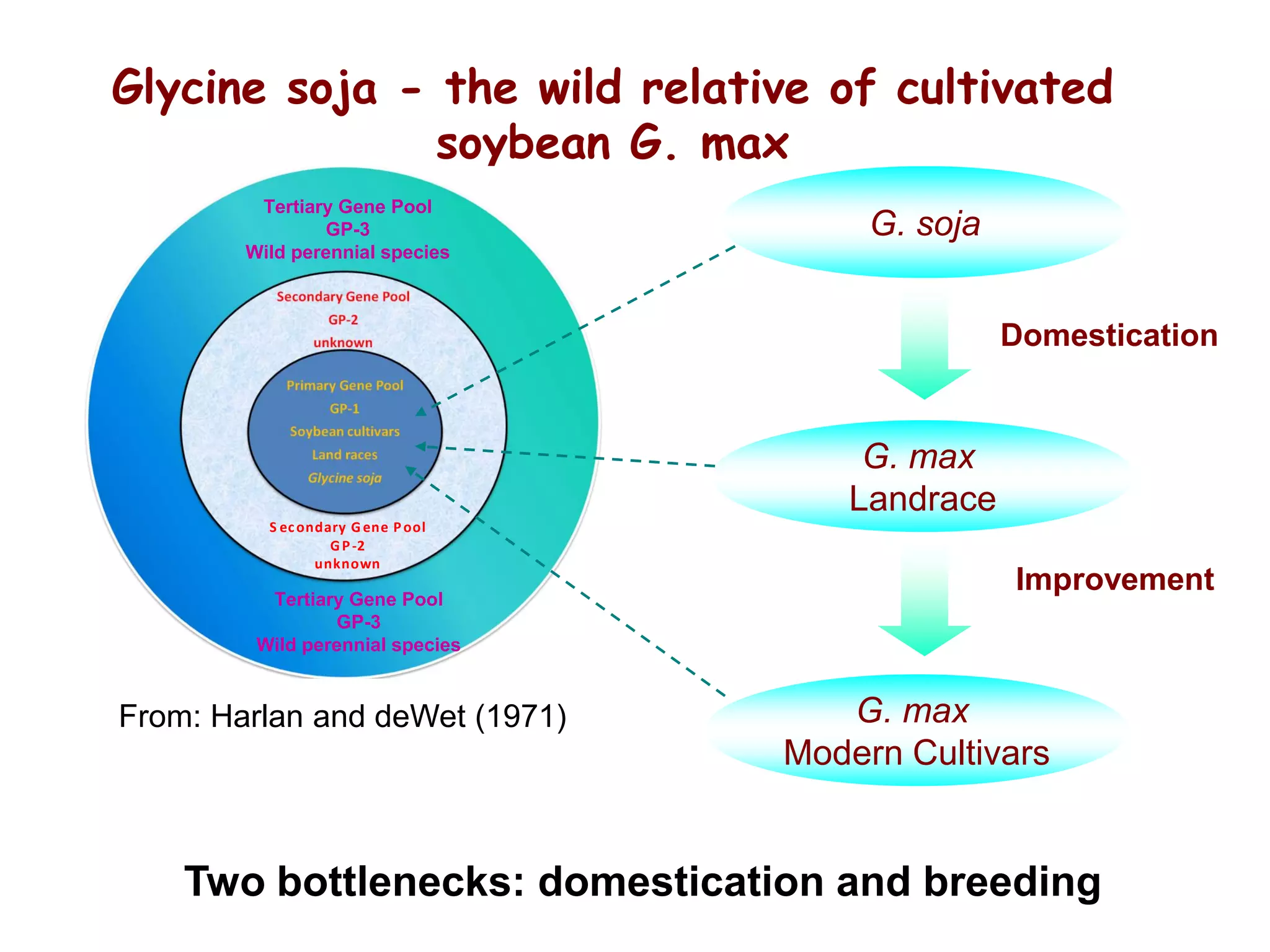
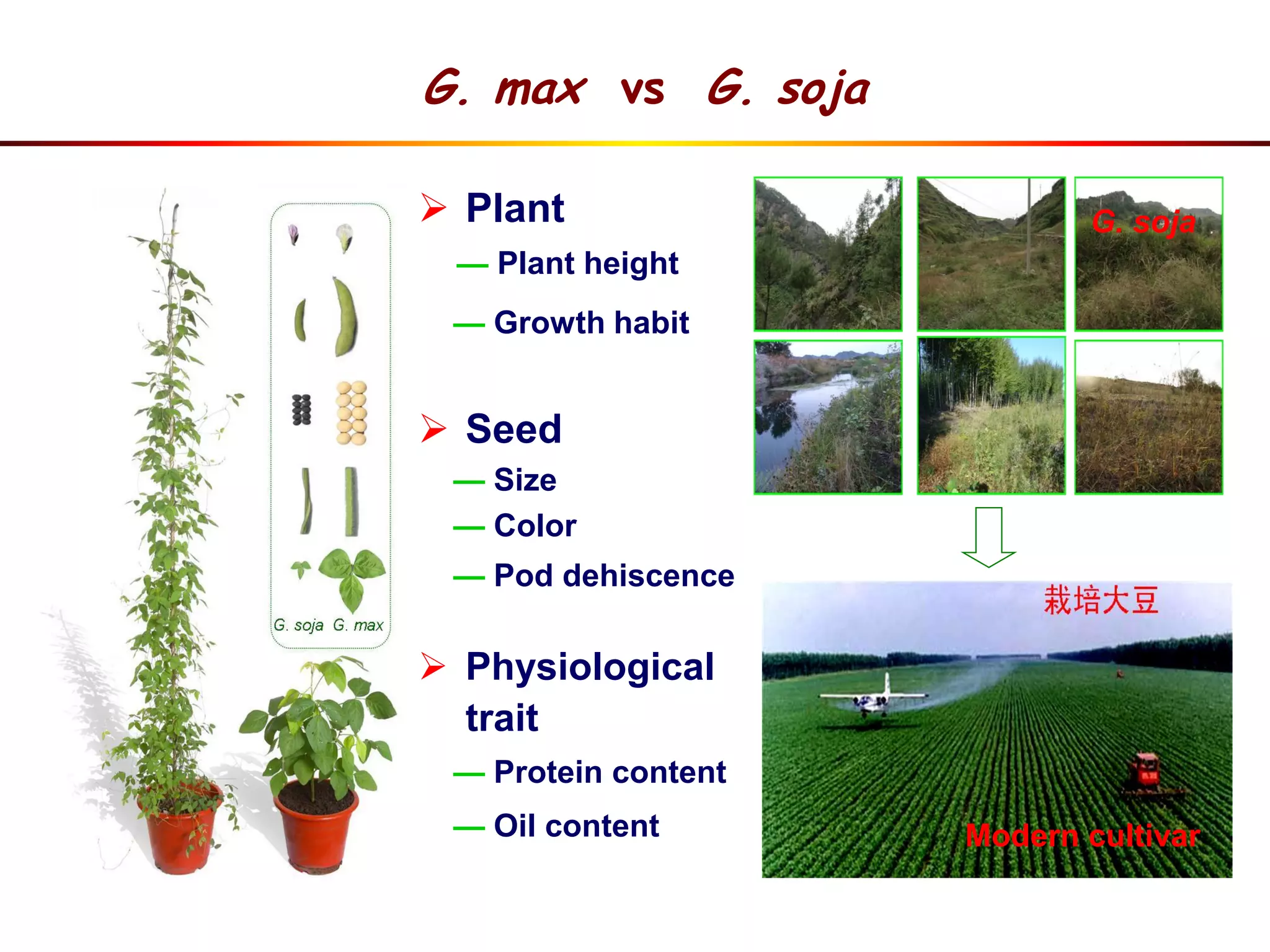
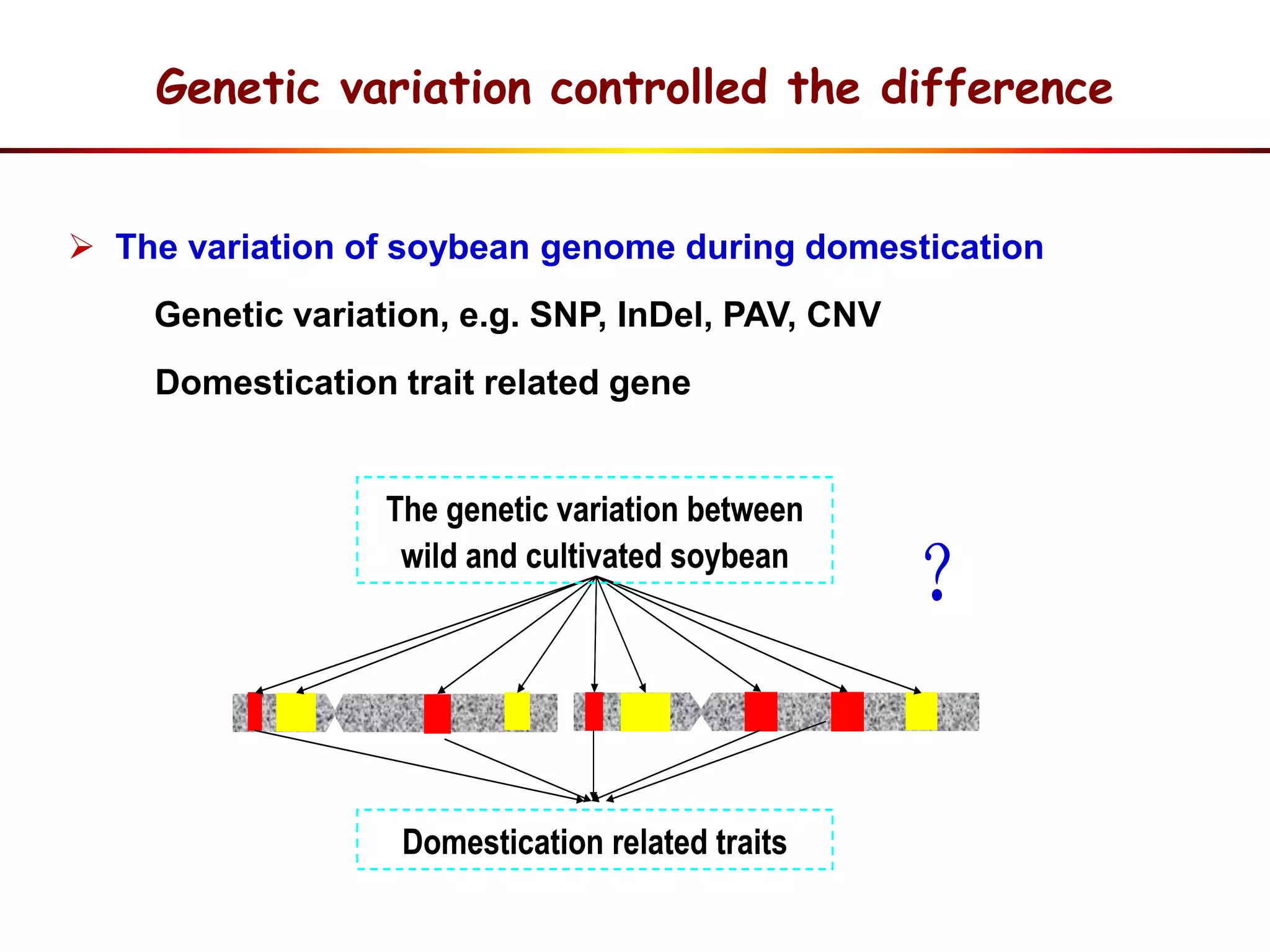
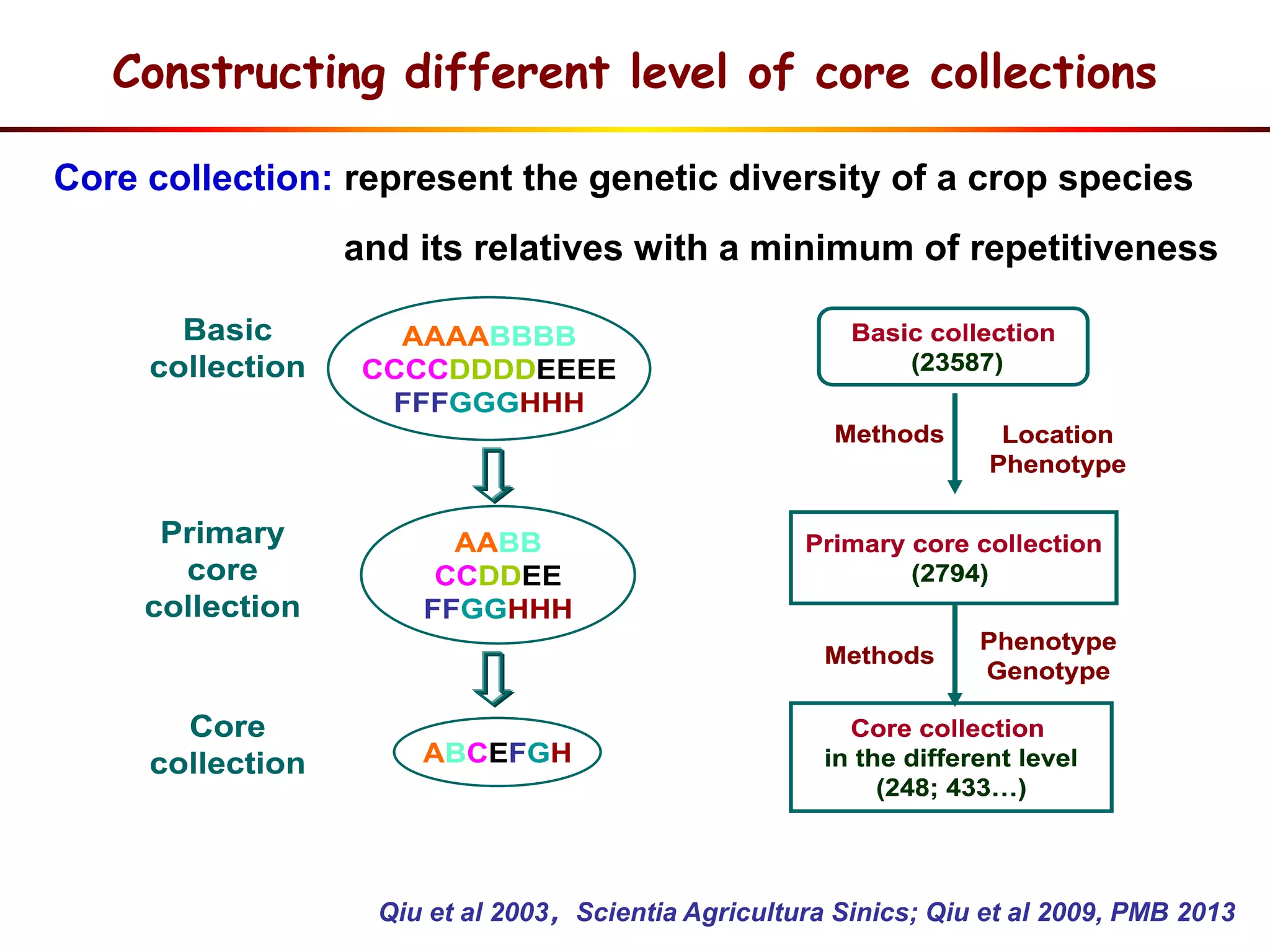
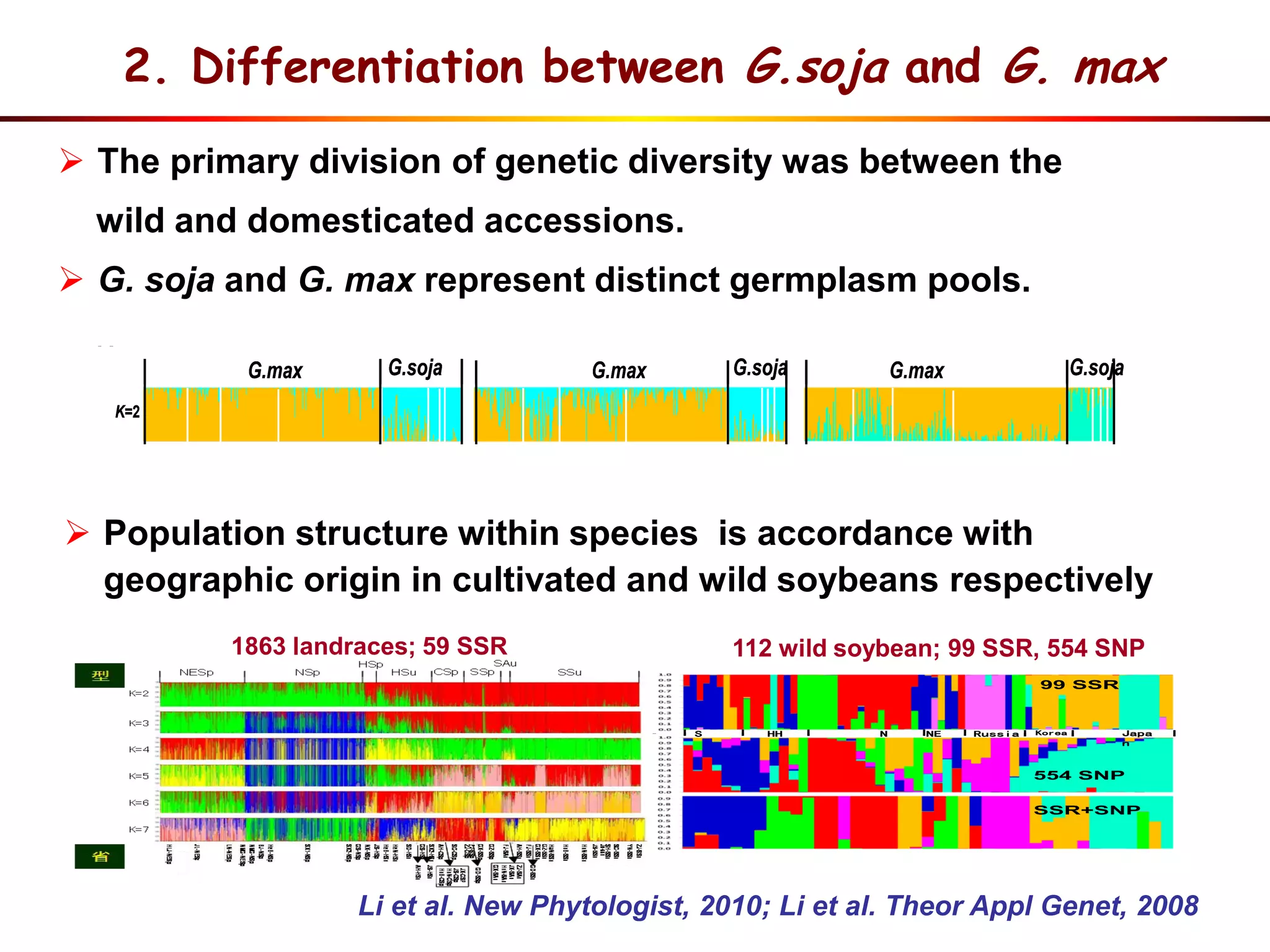
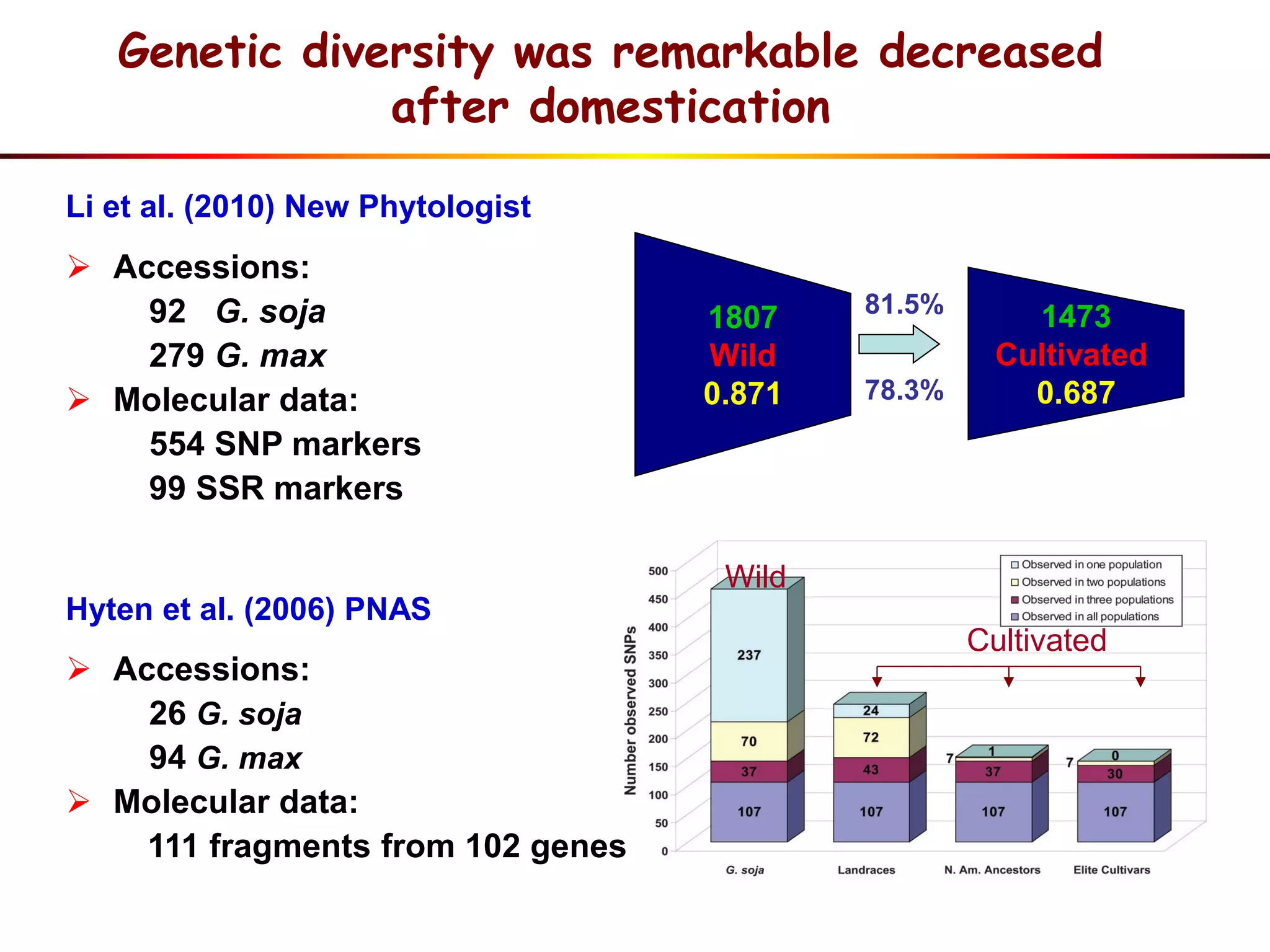
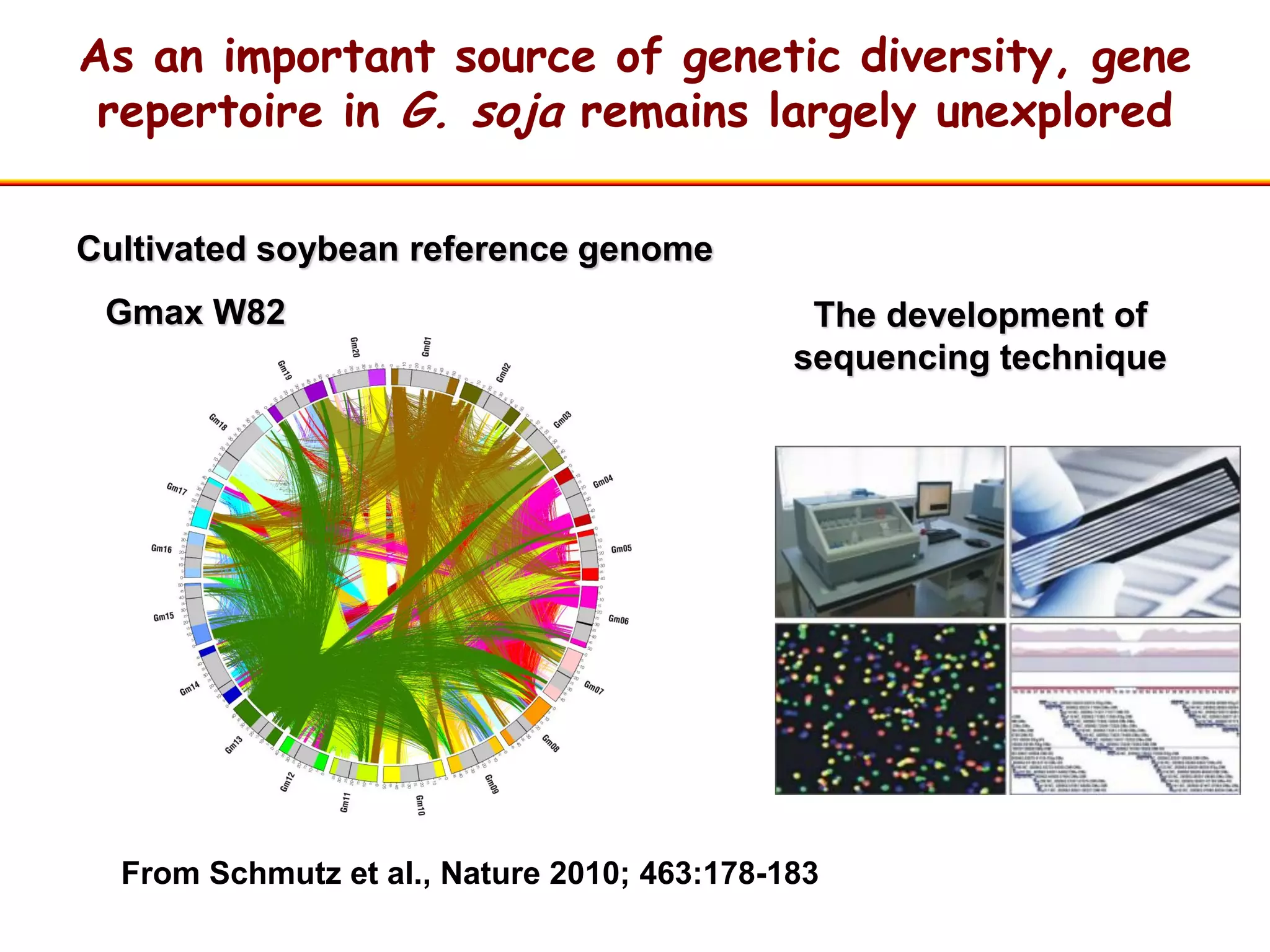
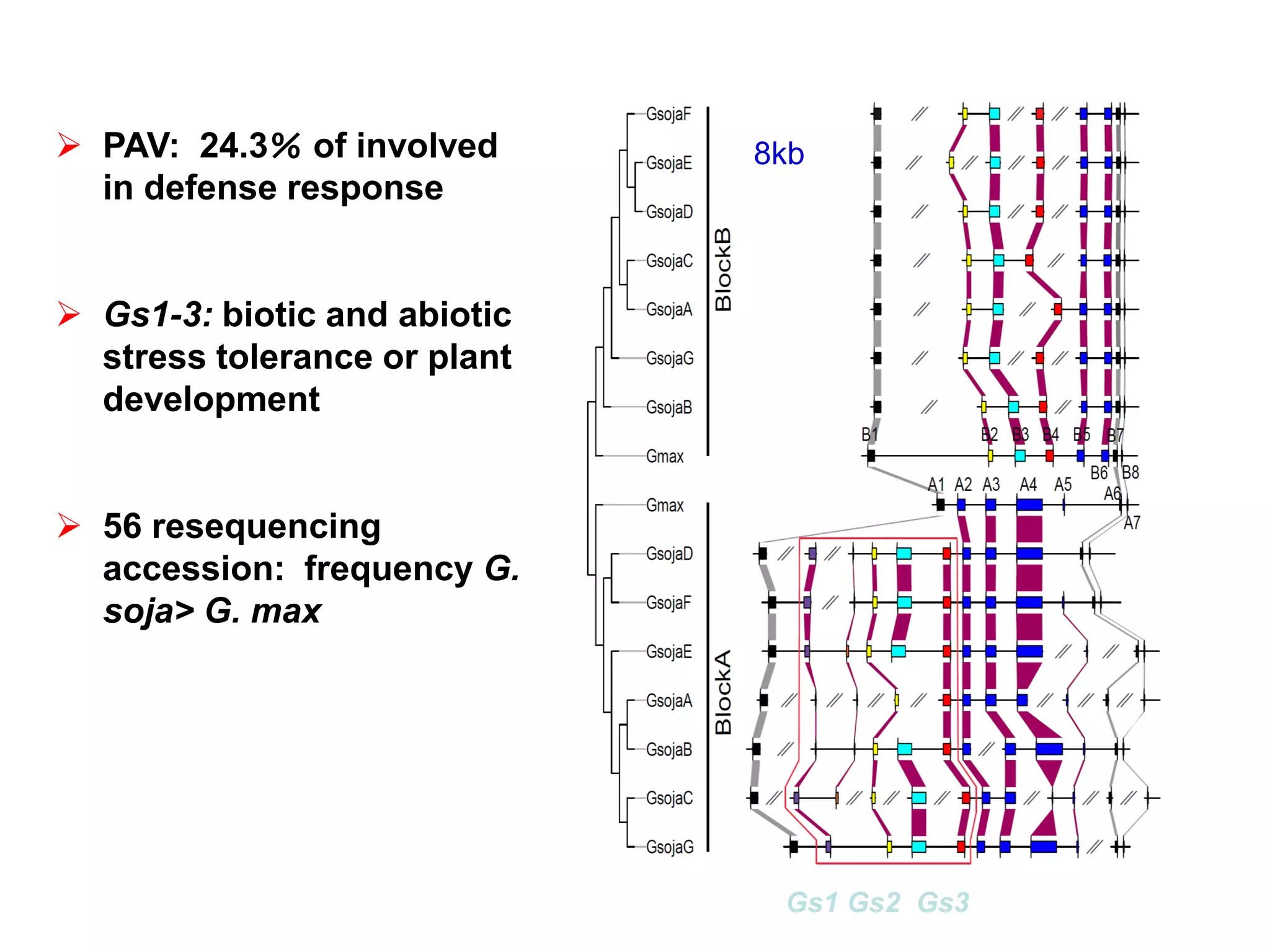
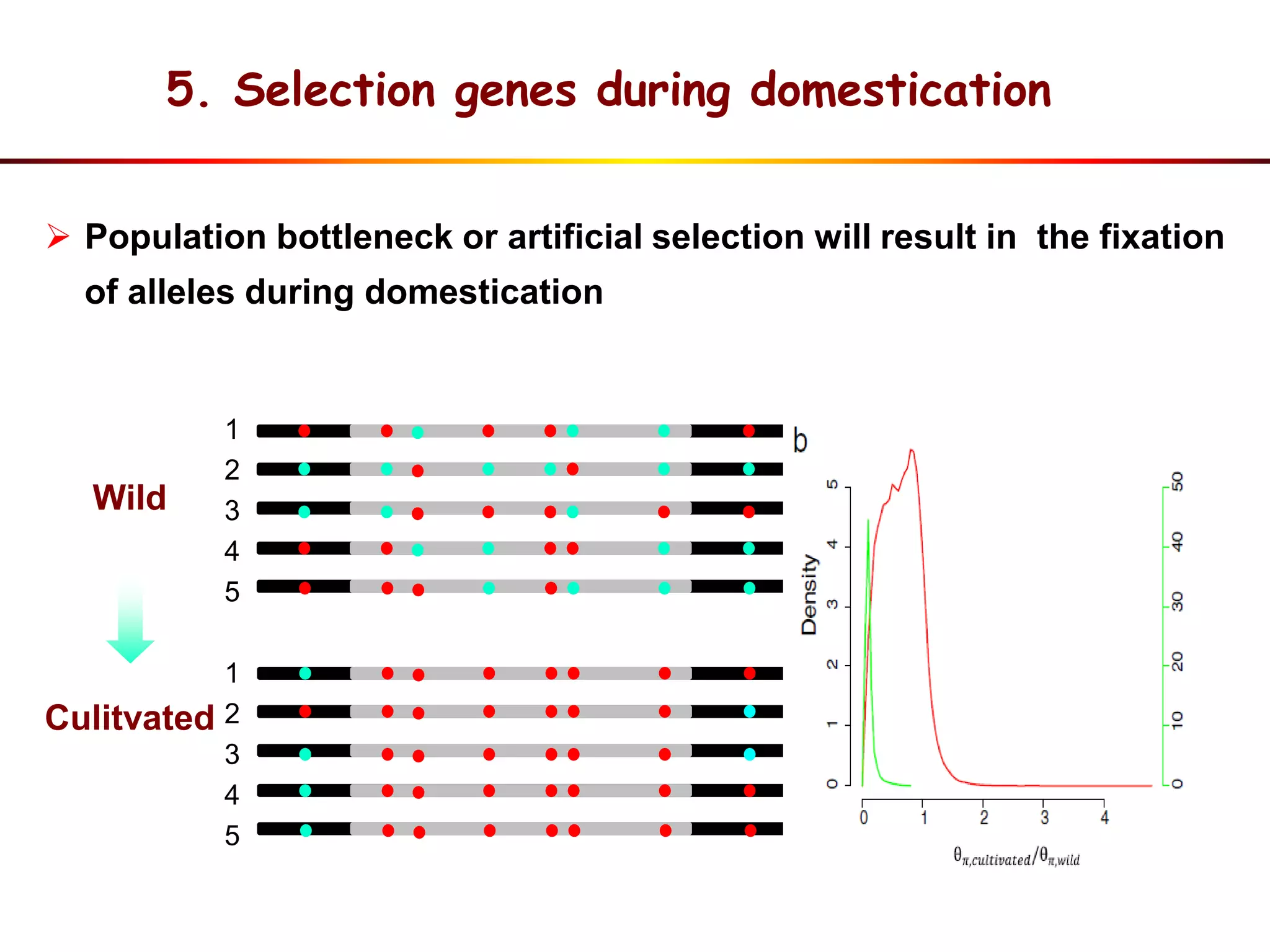
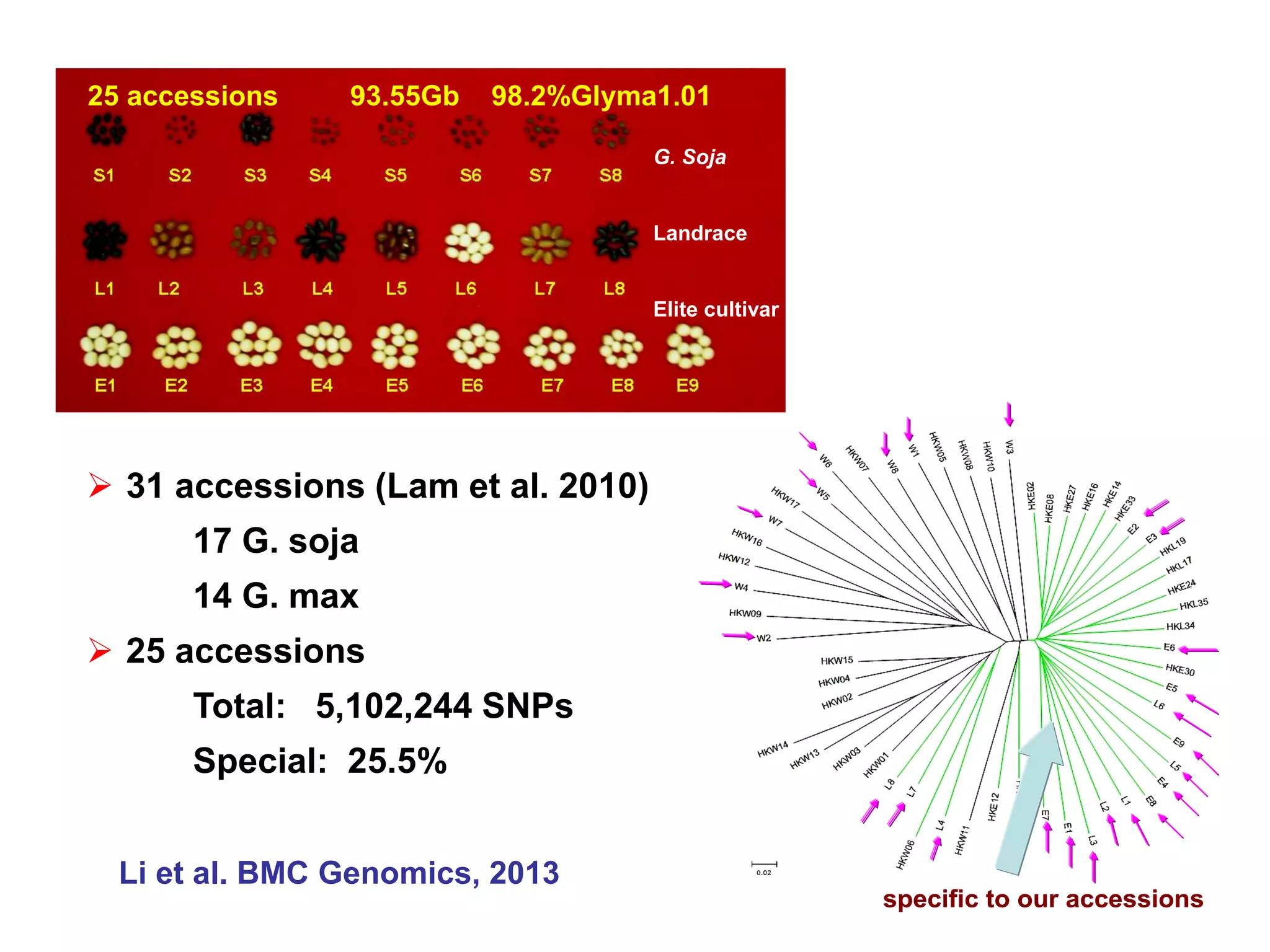
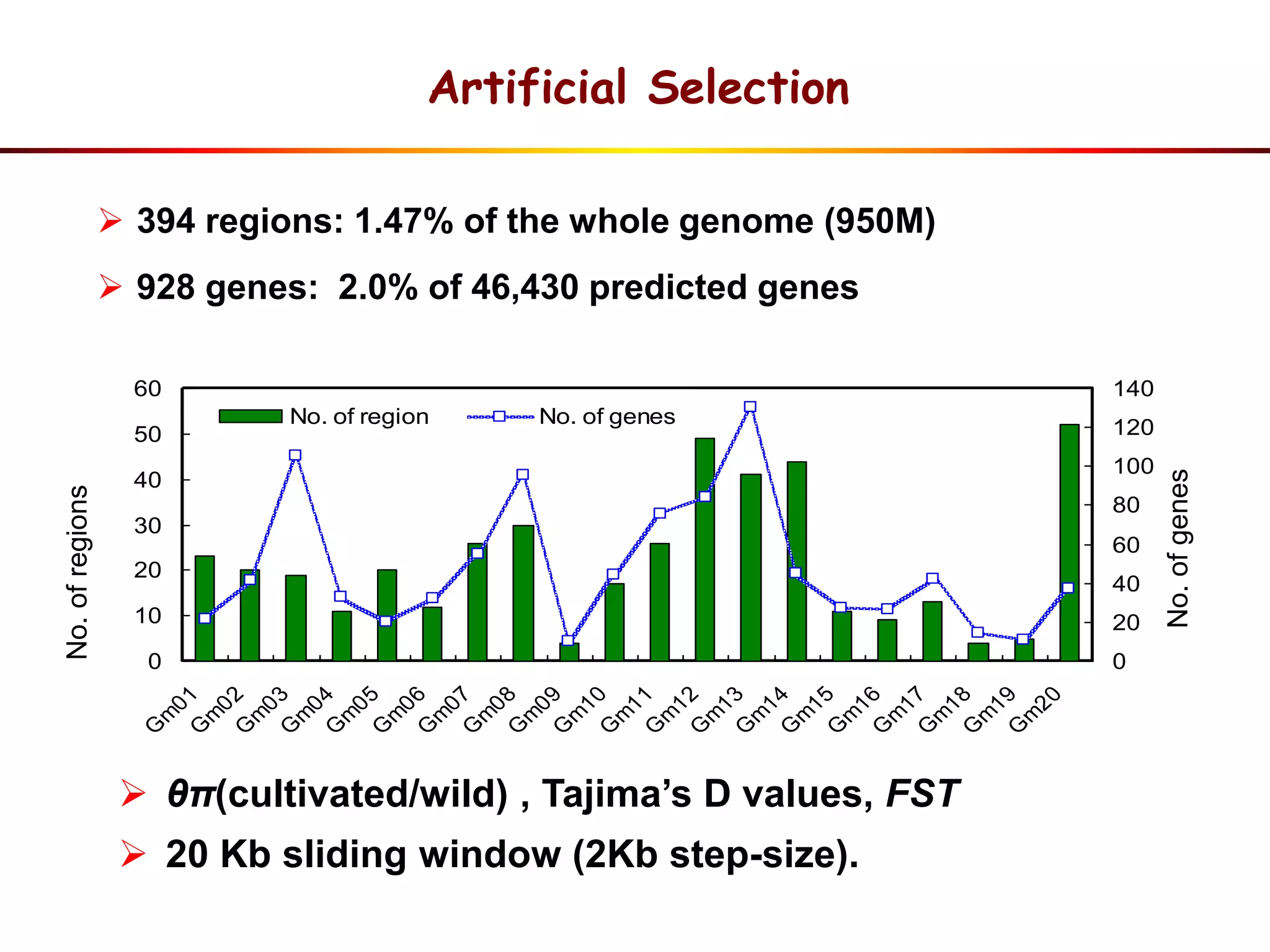
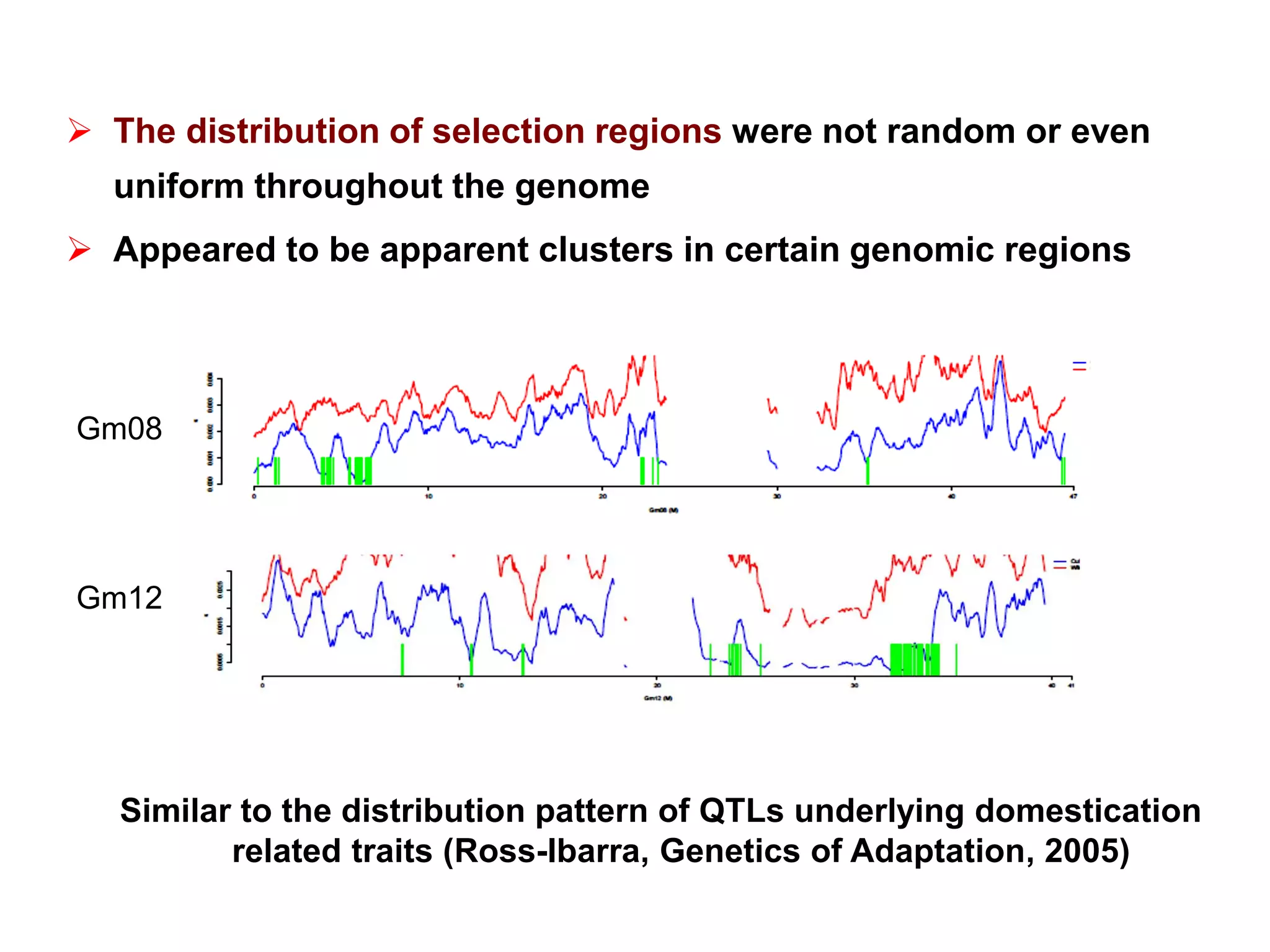
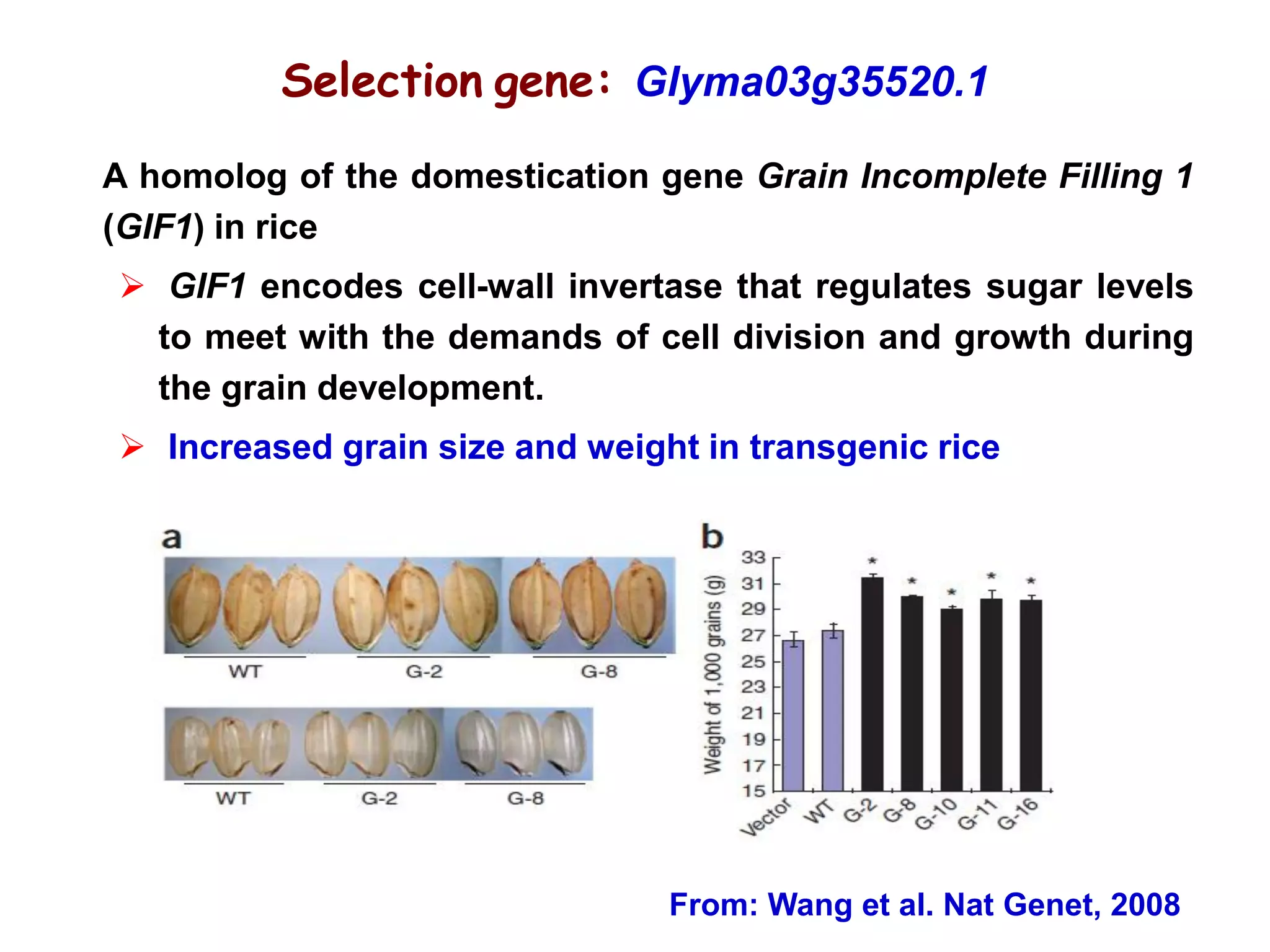
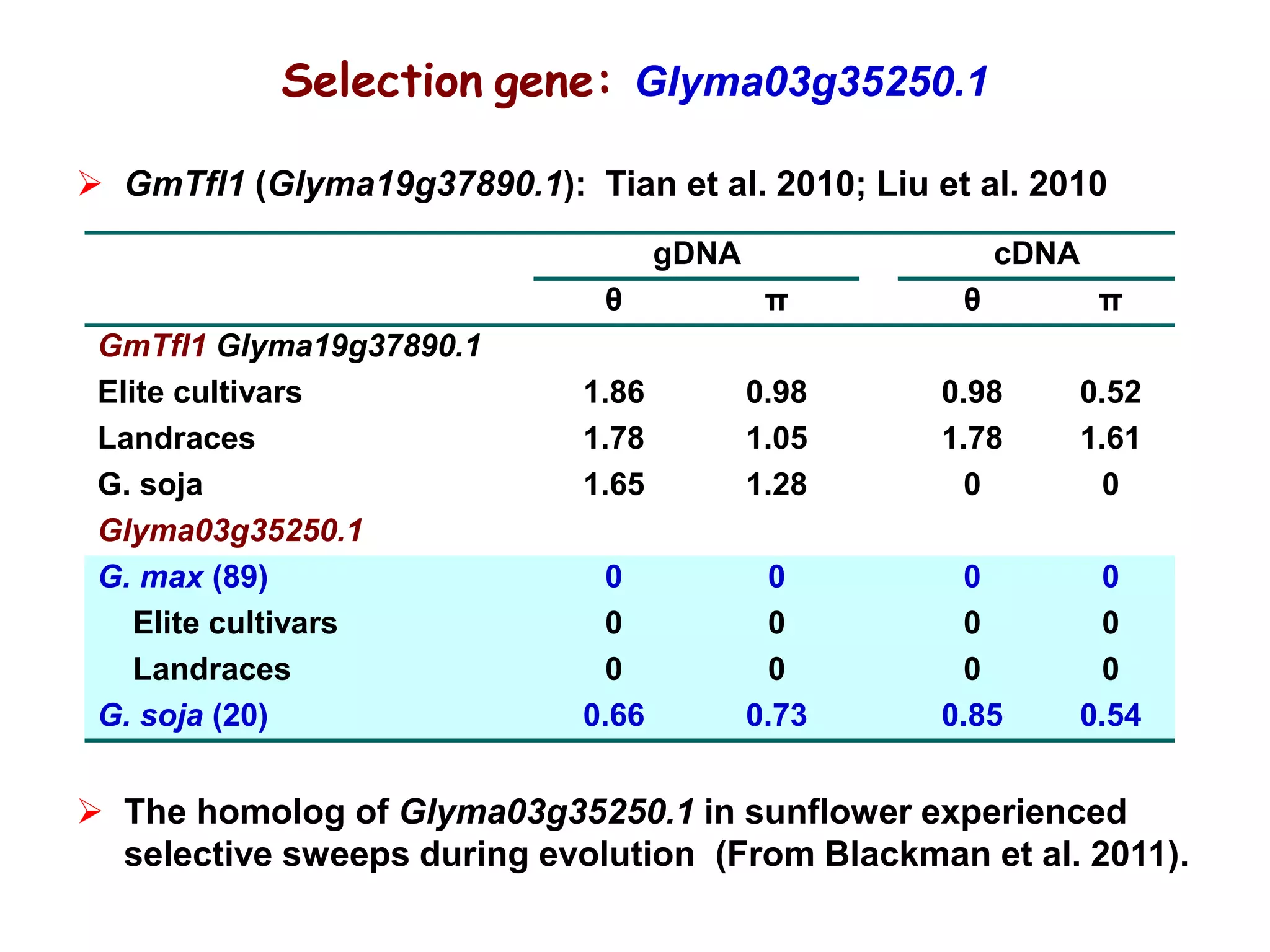
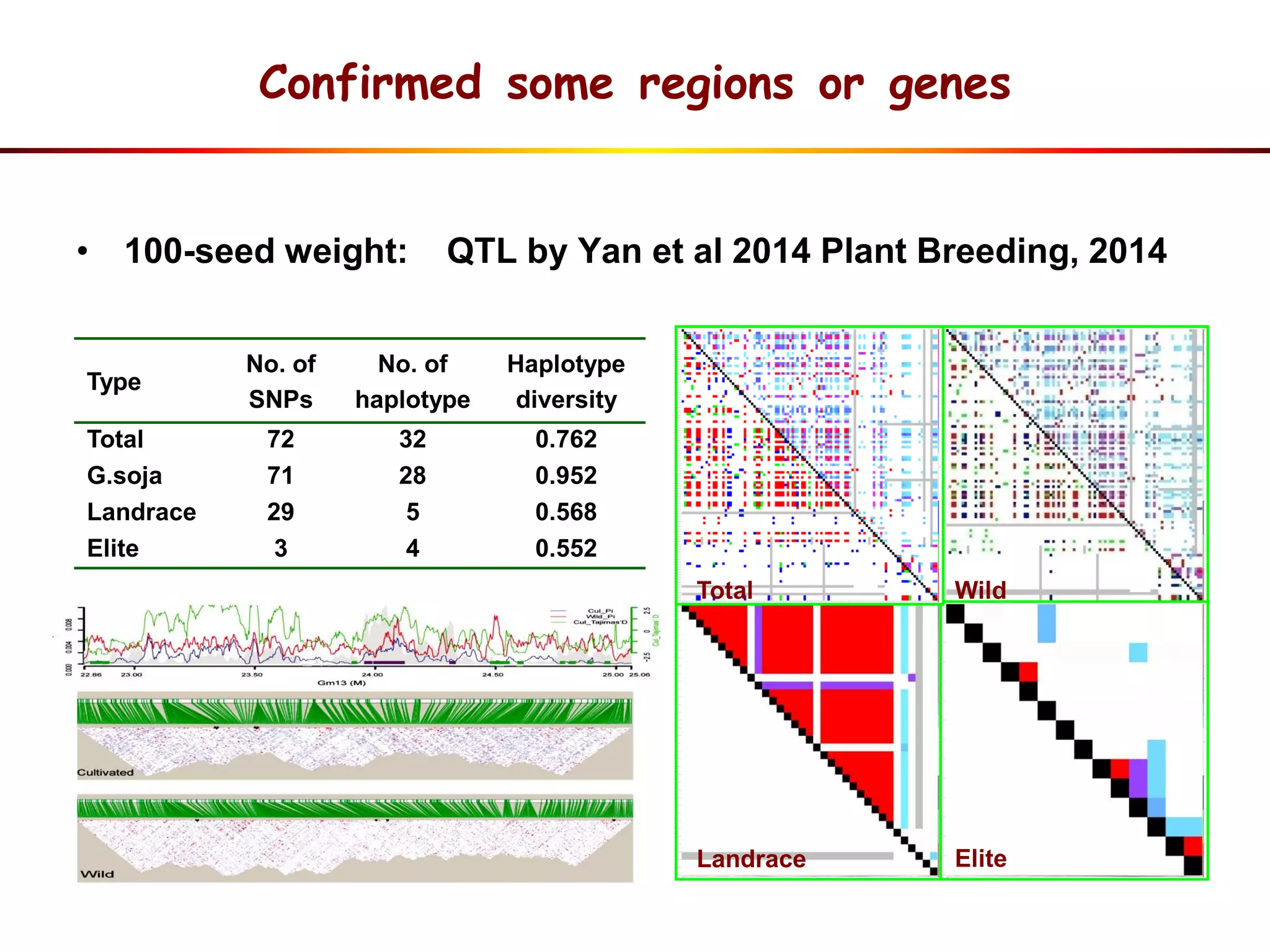
This document summarizes research on the genomic diversity of soybeans during domestication. It finds:
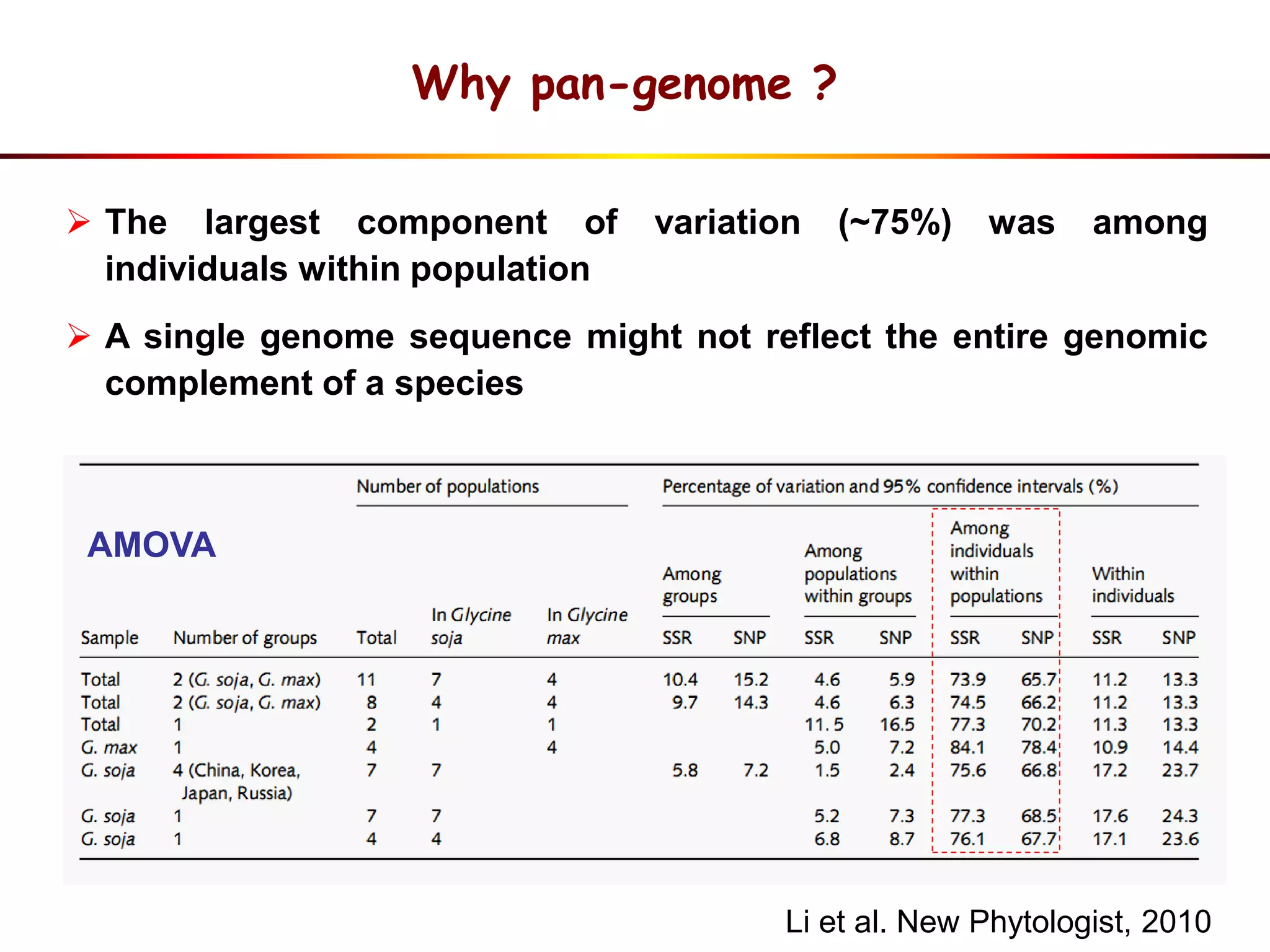
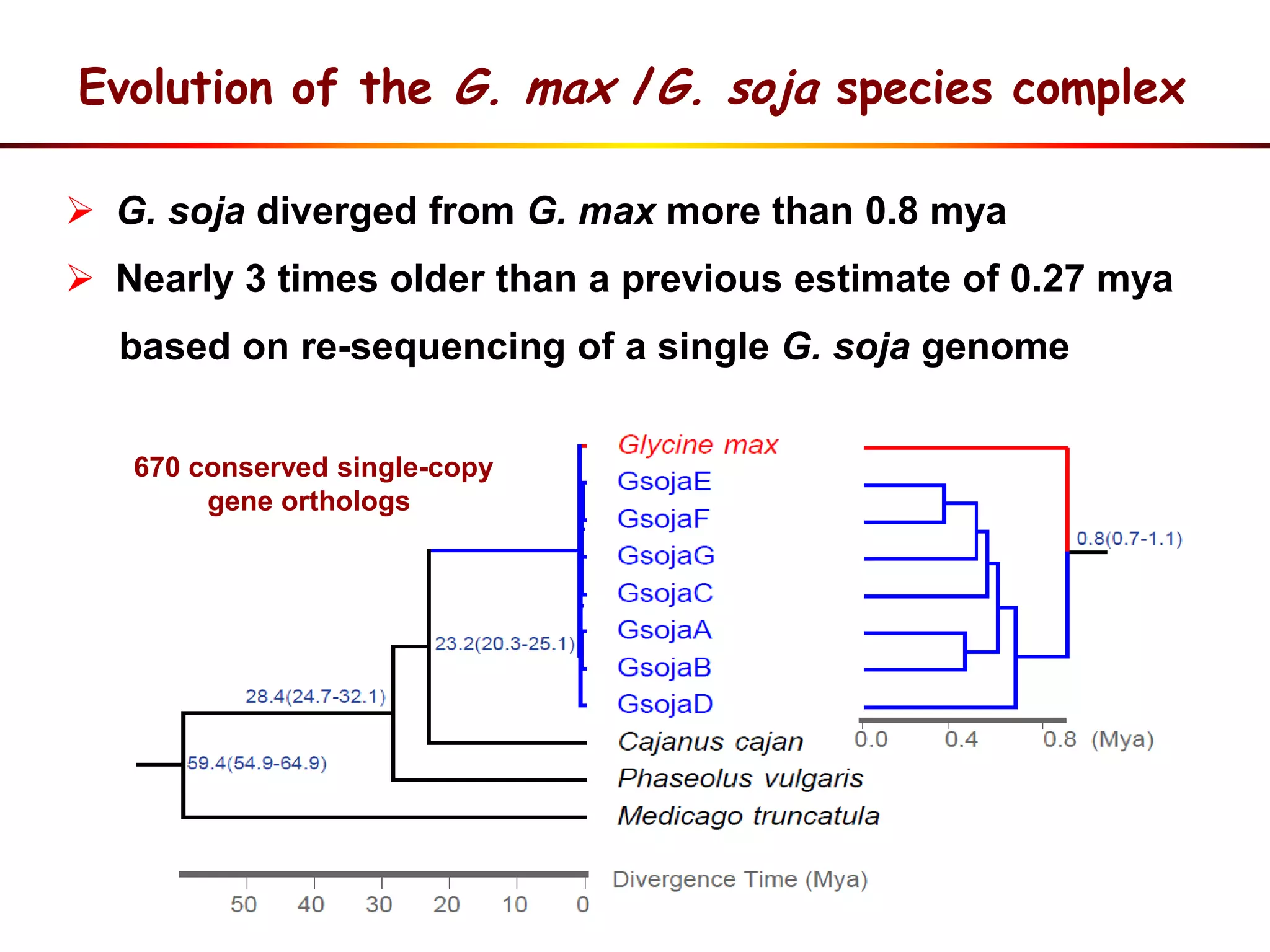
1) Wild soybeans (G. soja) show more genetic diversity than cultivated soybeans (G. max) due to bottlenecks during domestication and breeding.
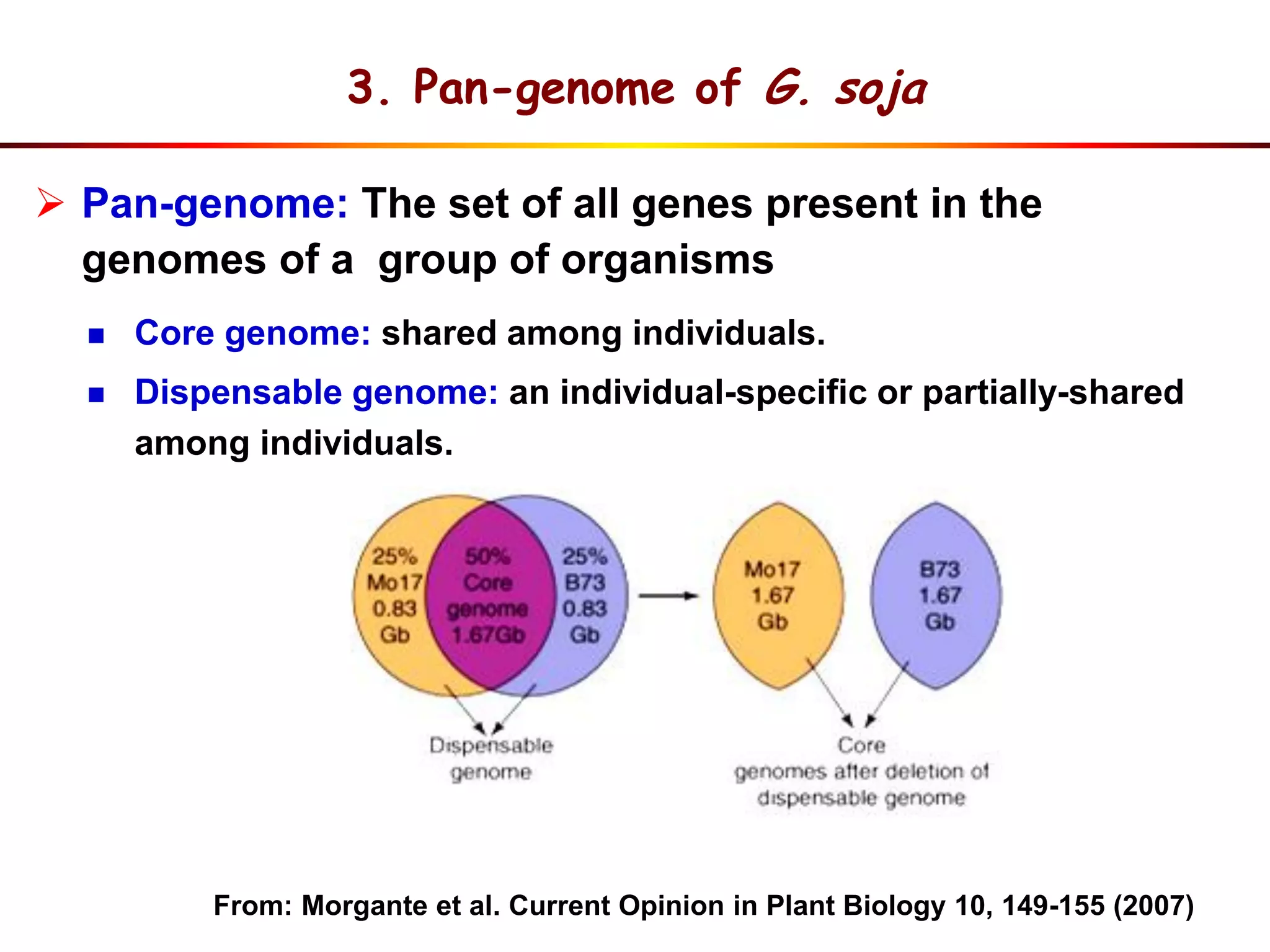
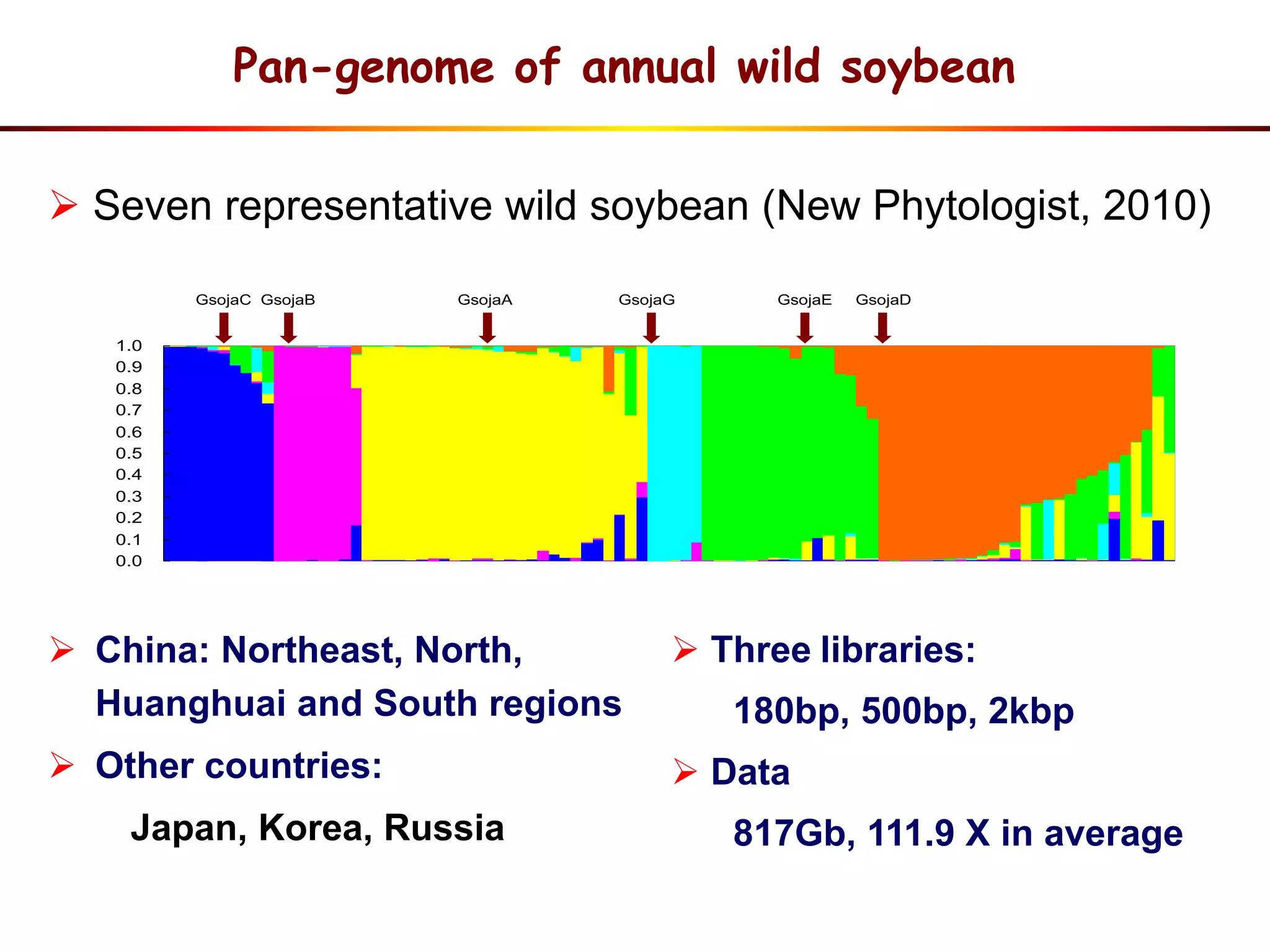
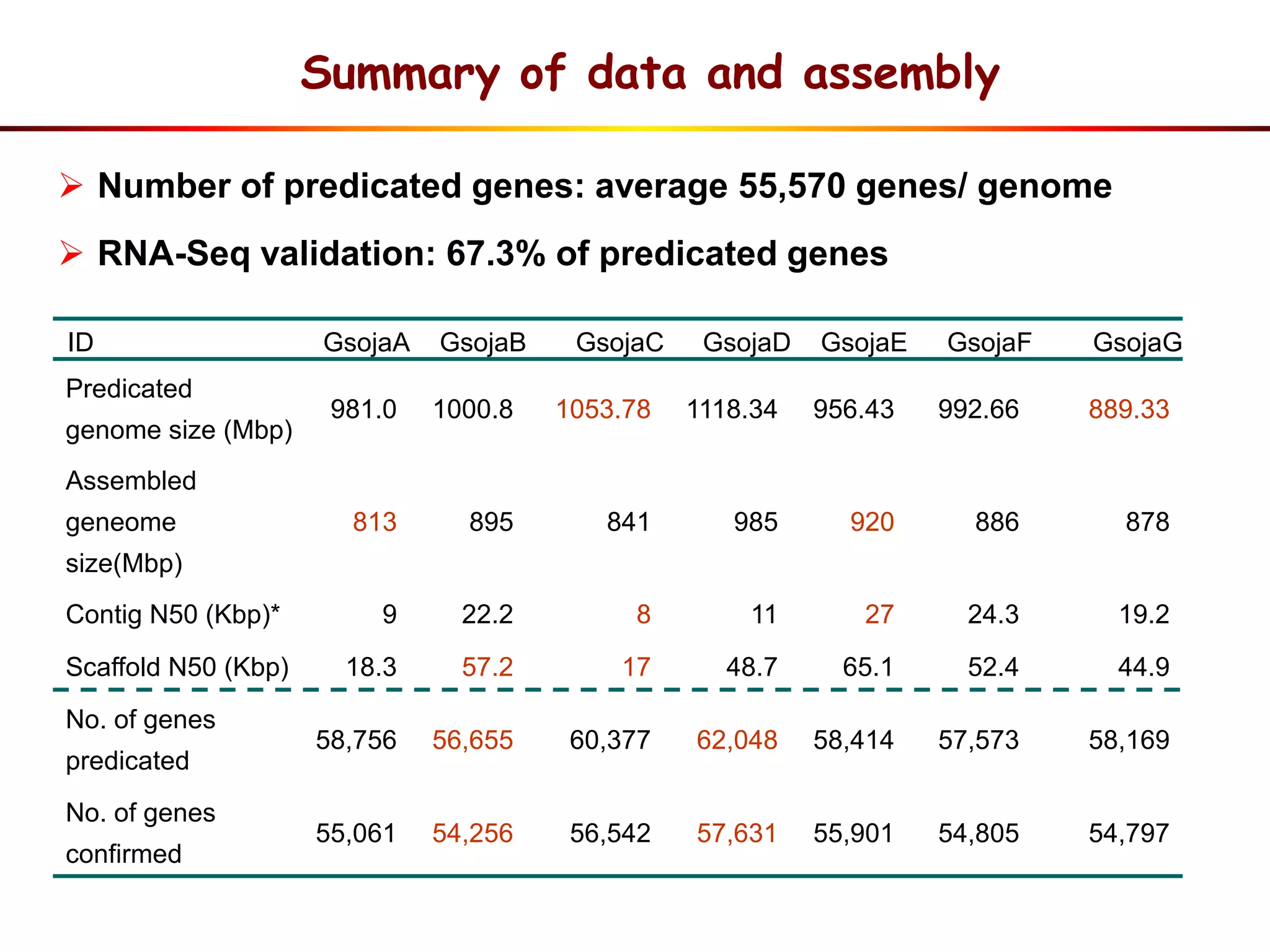
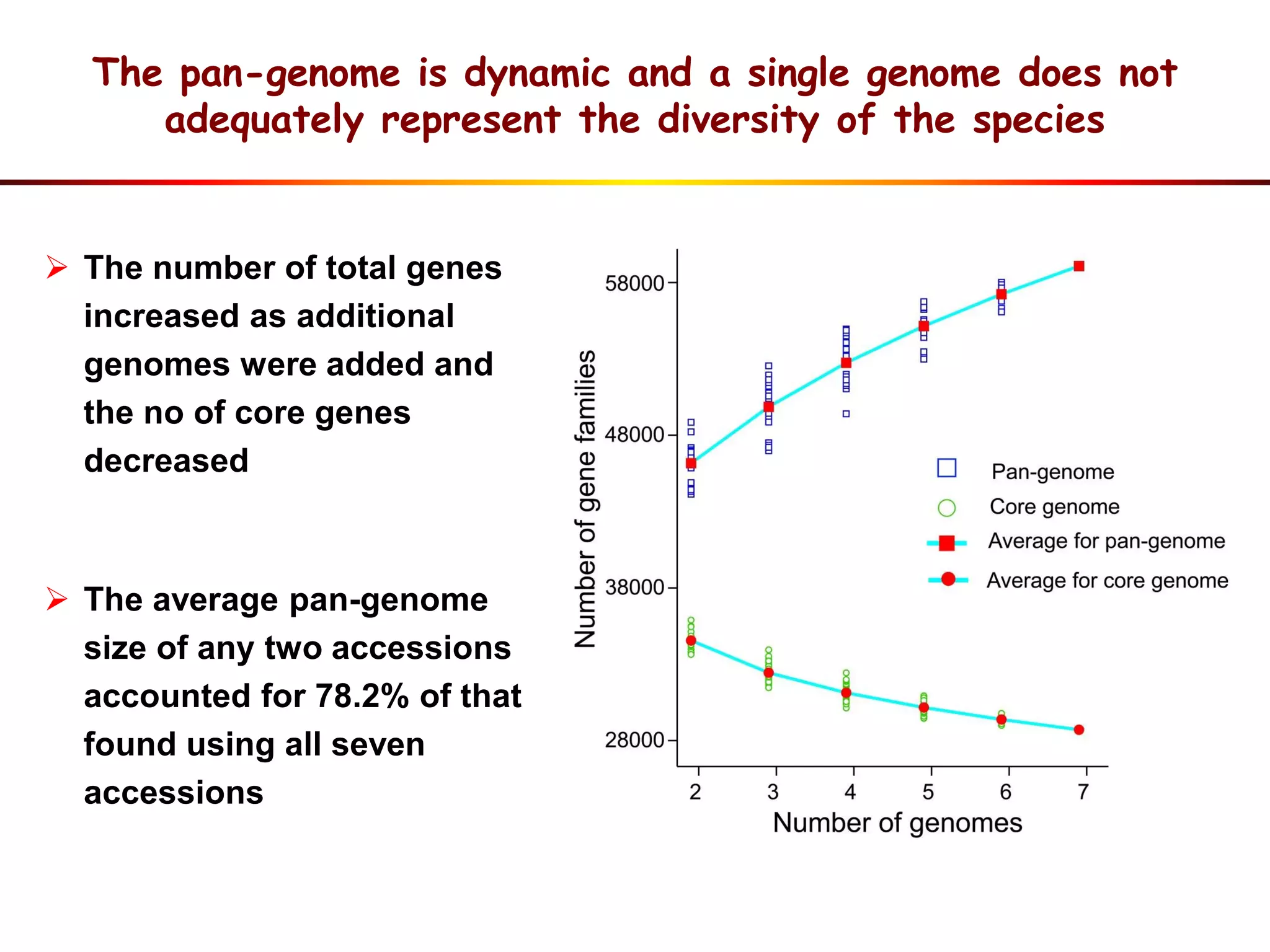
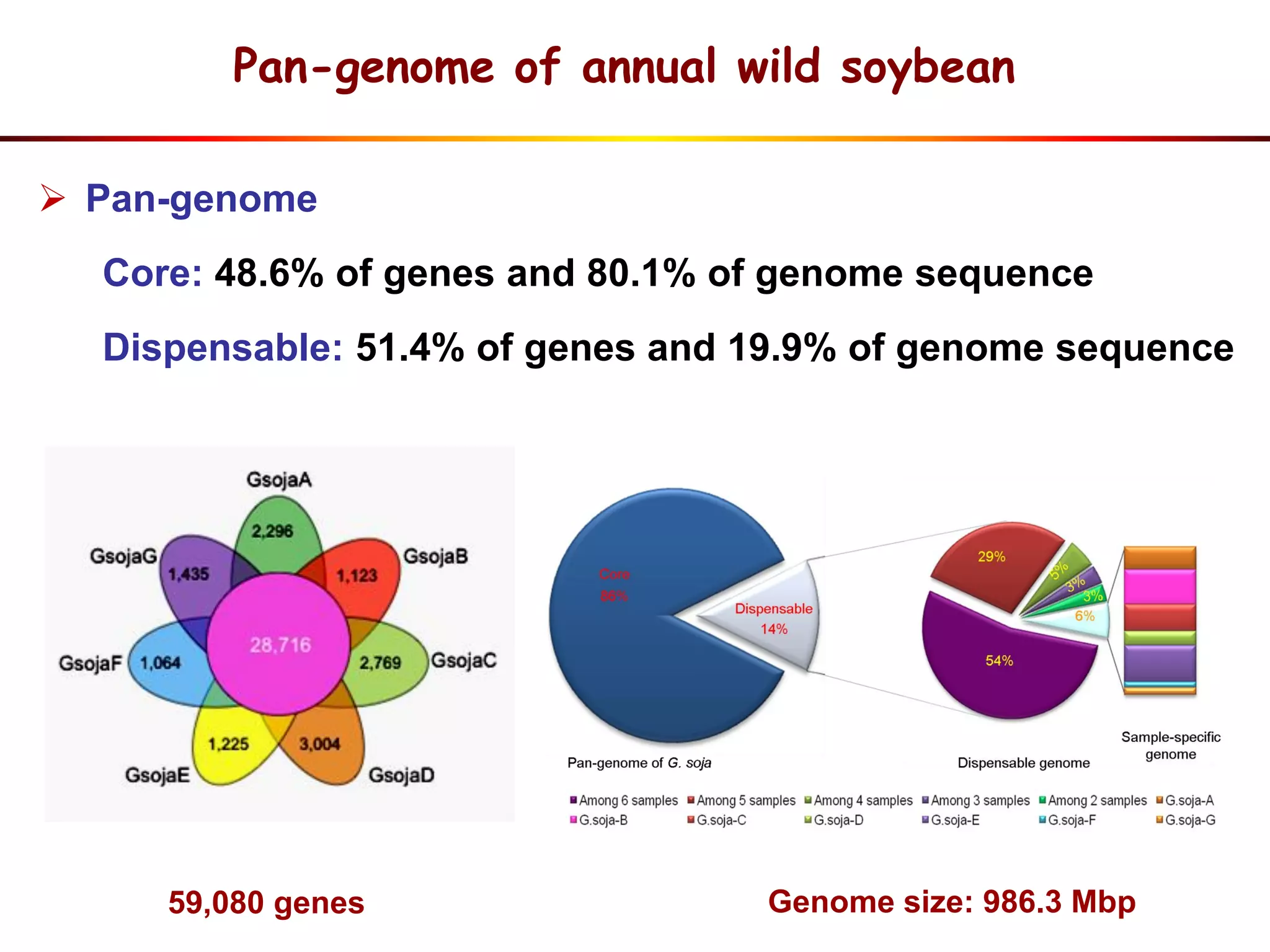
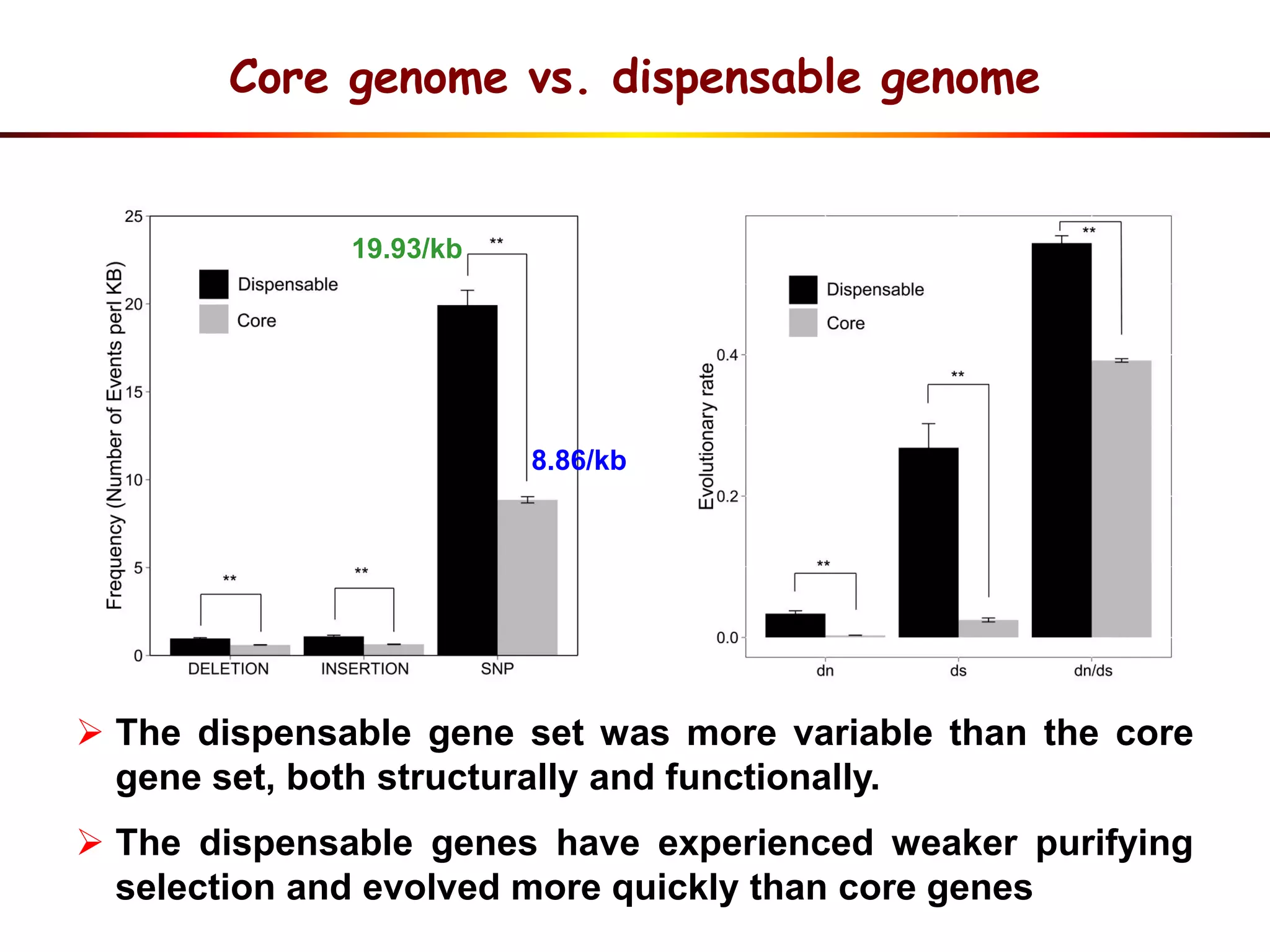
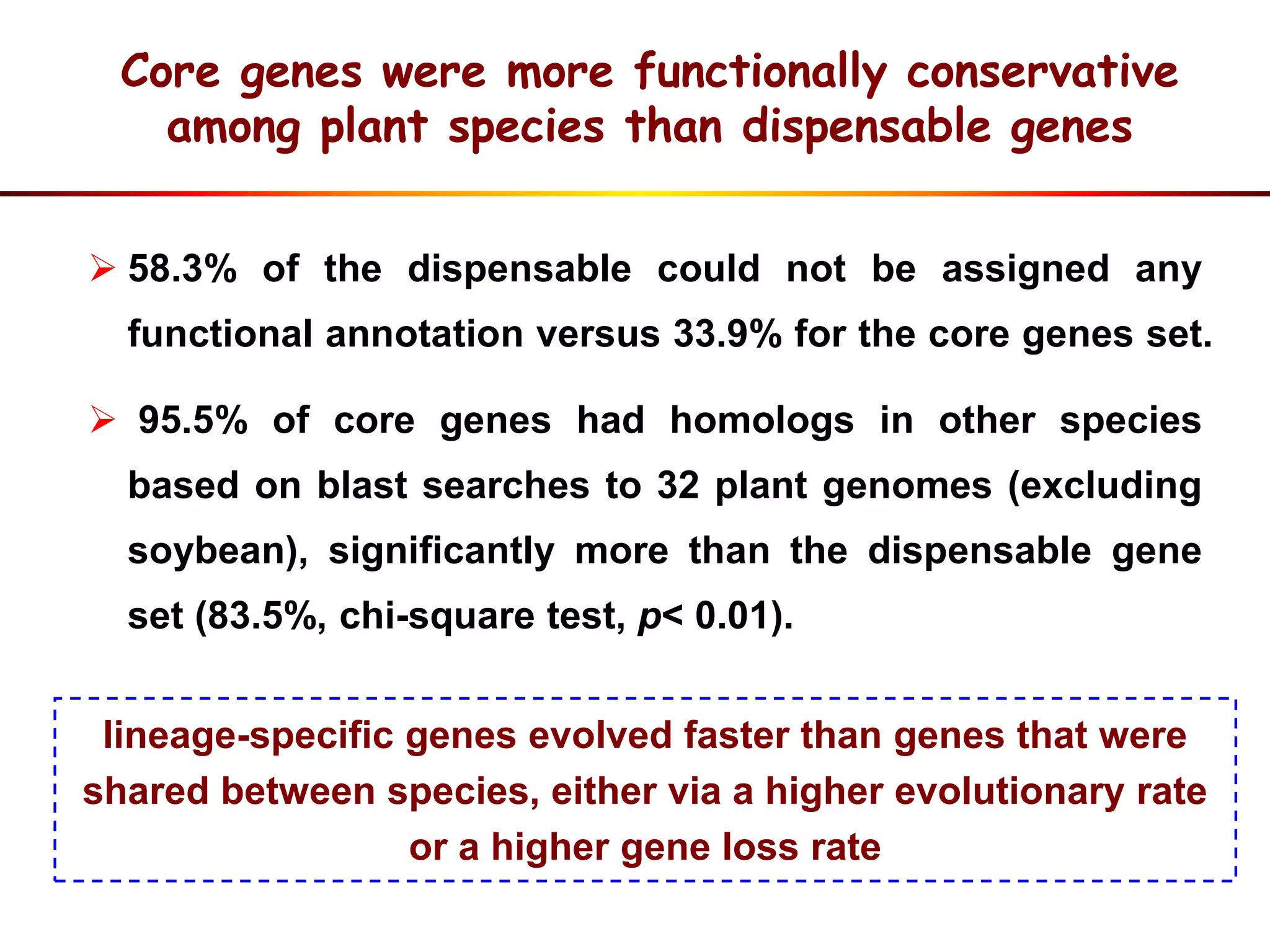
2) Analysis of 7 G. soja genomes reveals a pan-genome structure with a large dispensable/variable gene set involved in environmental responses.
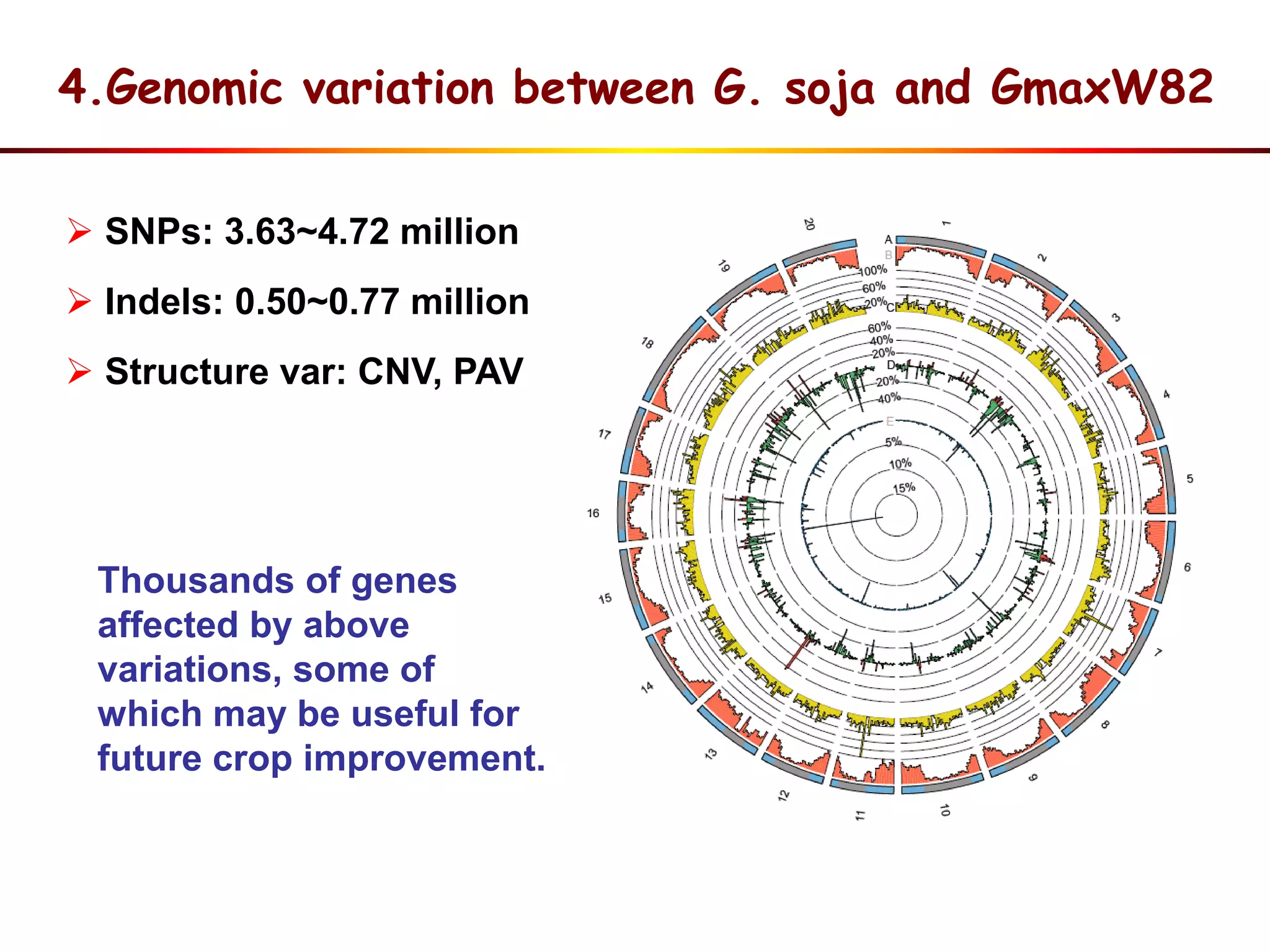
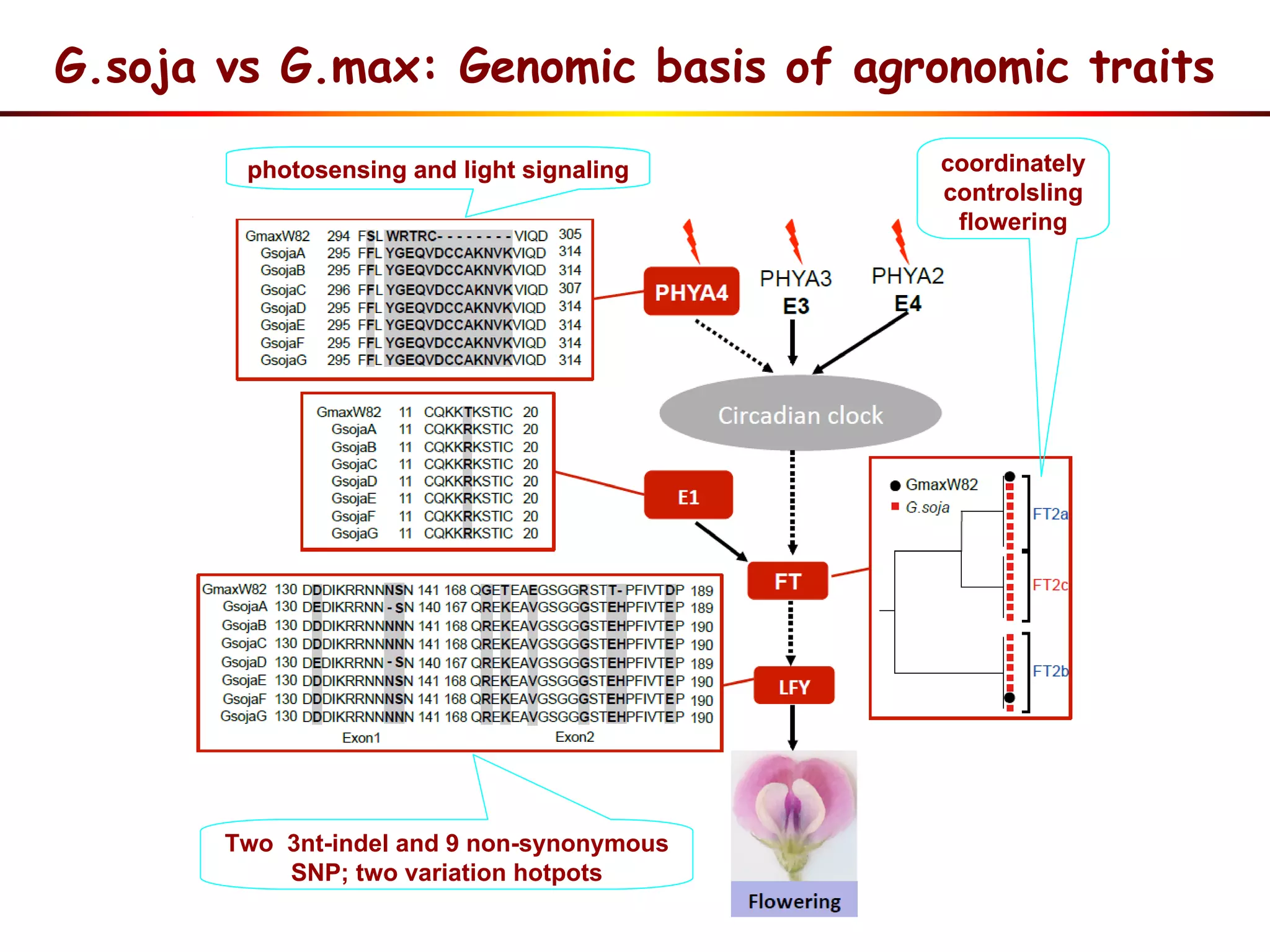
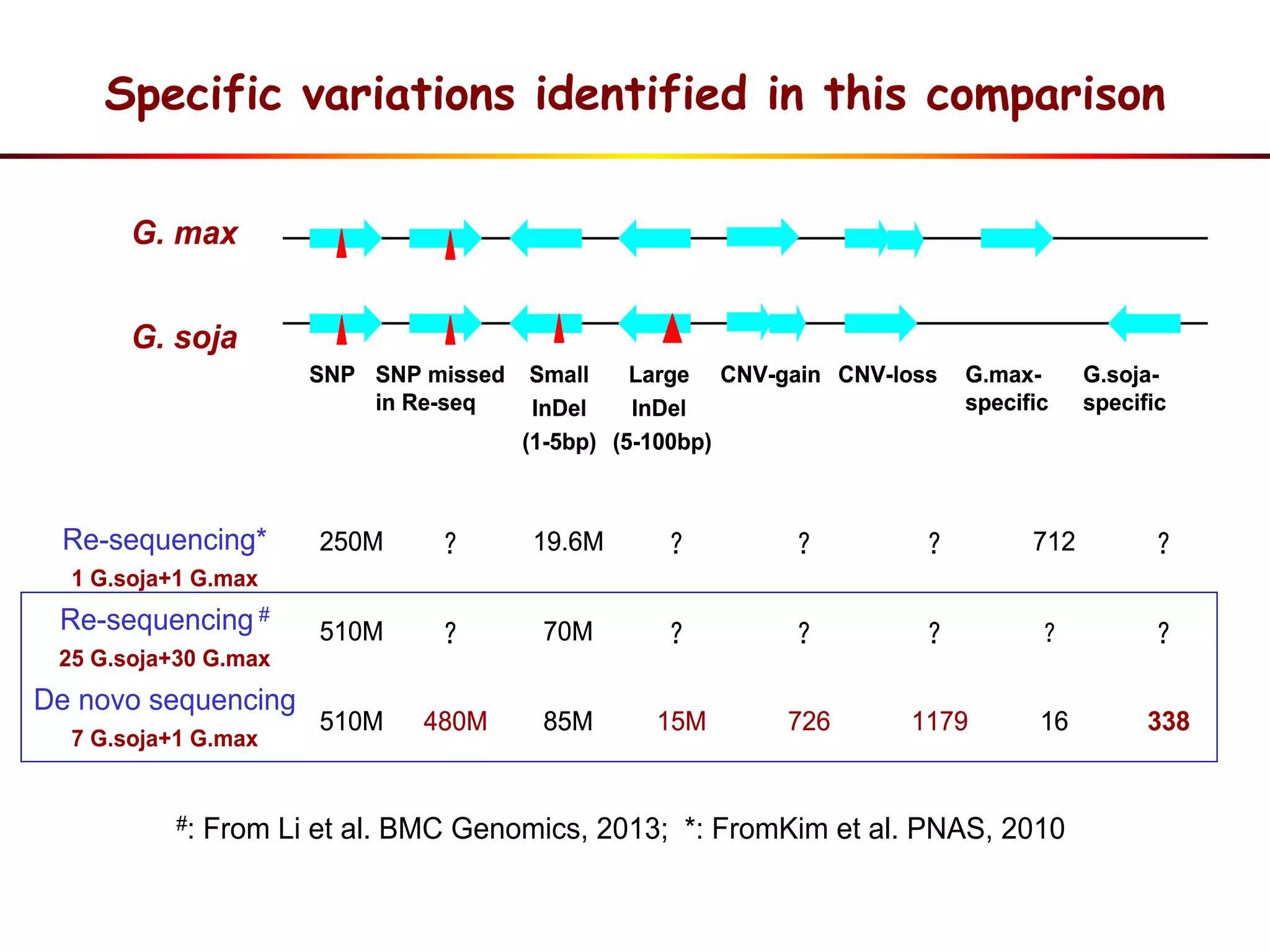
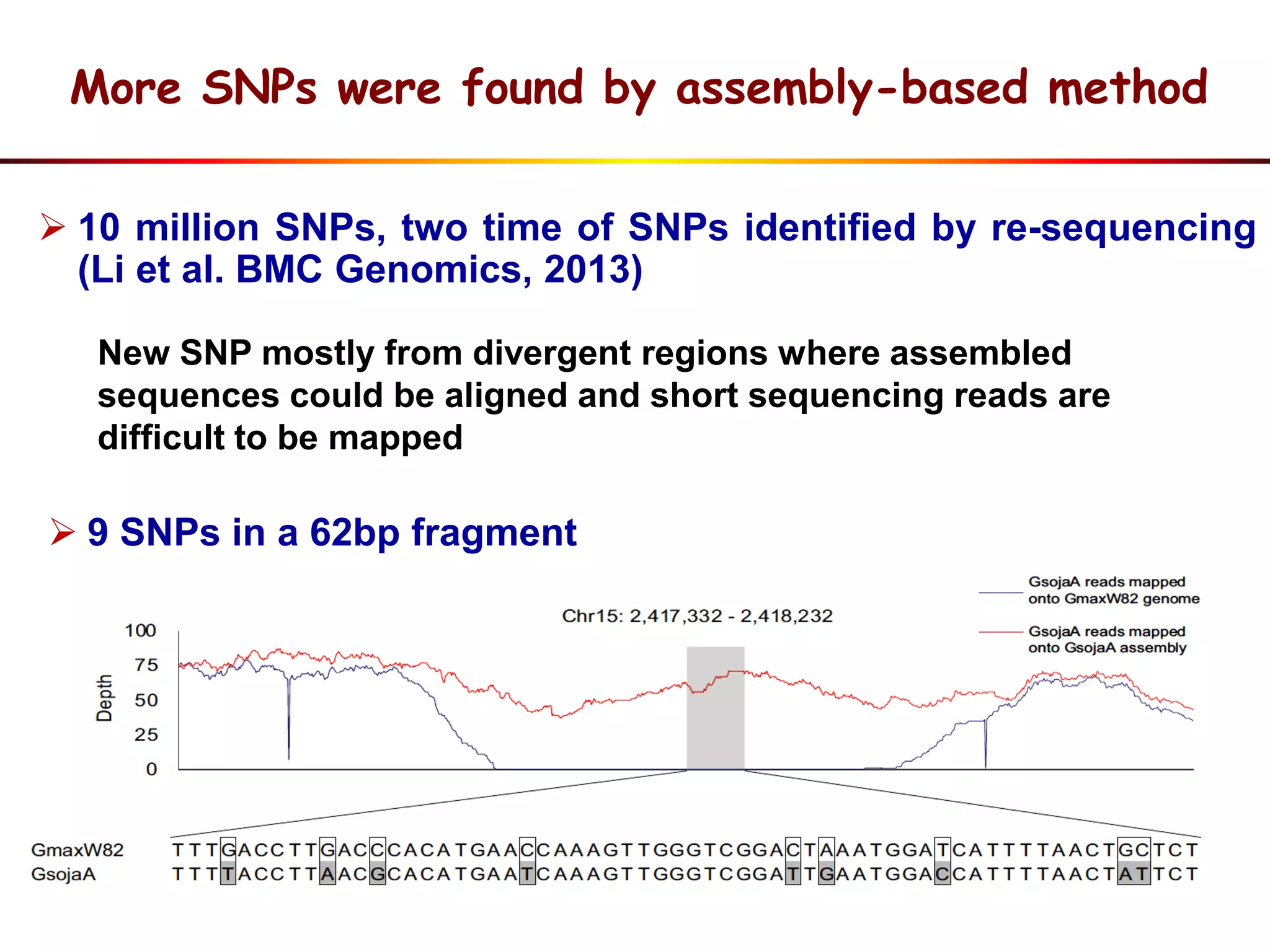
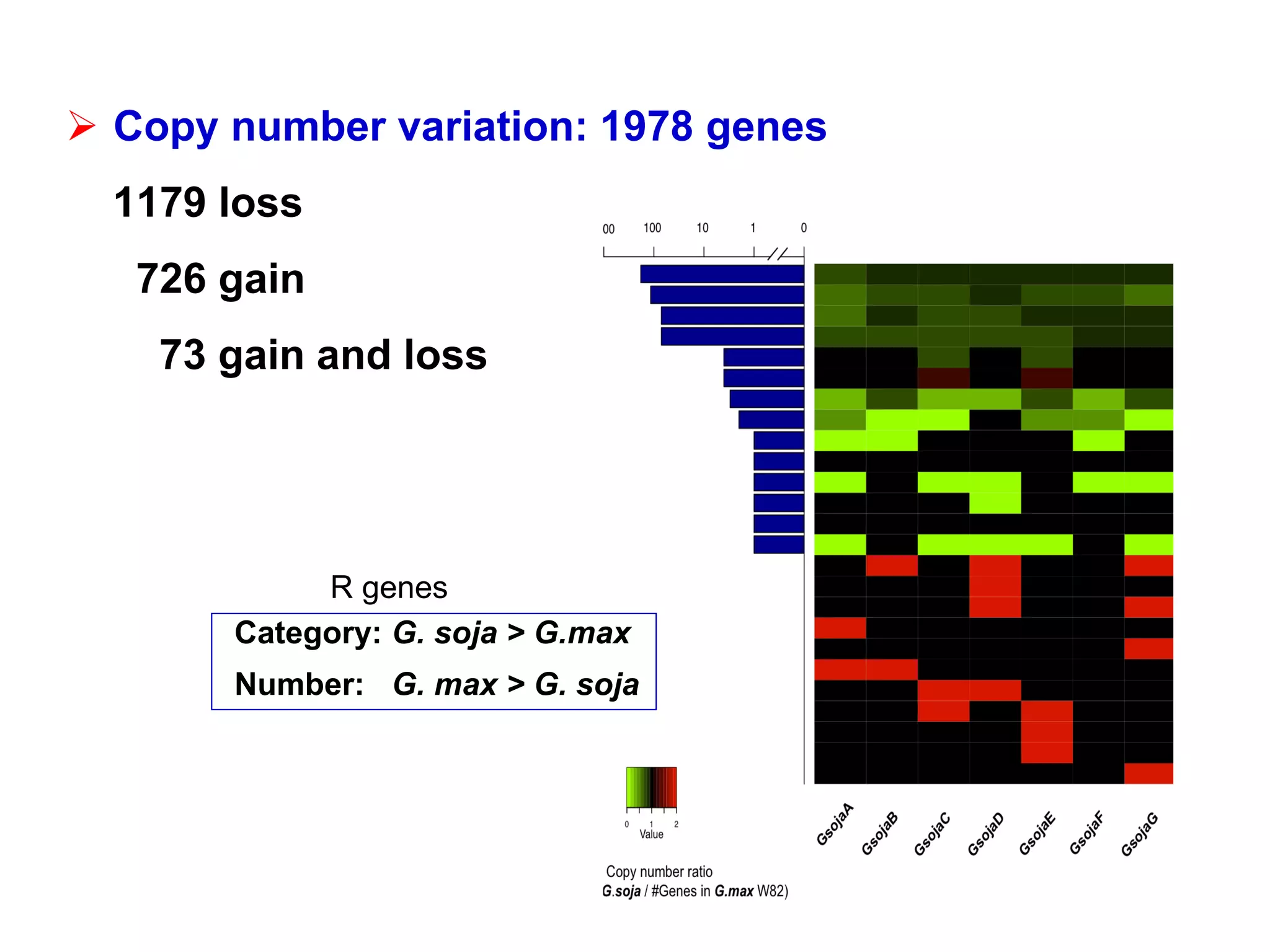
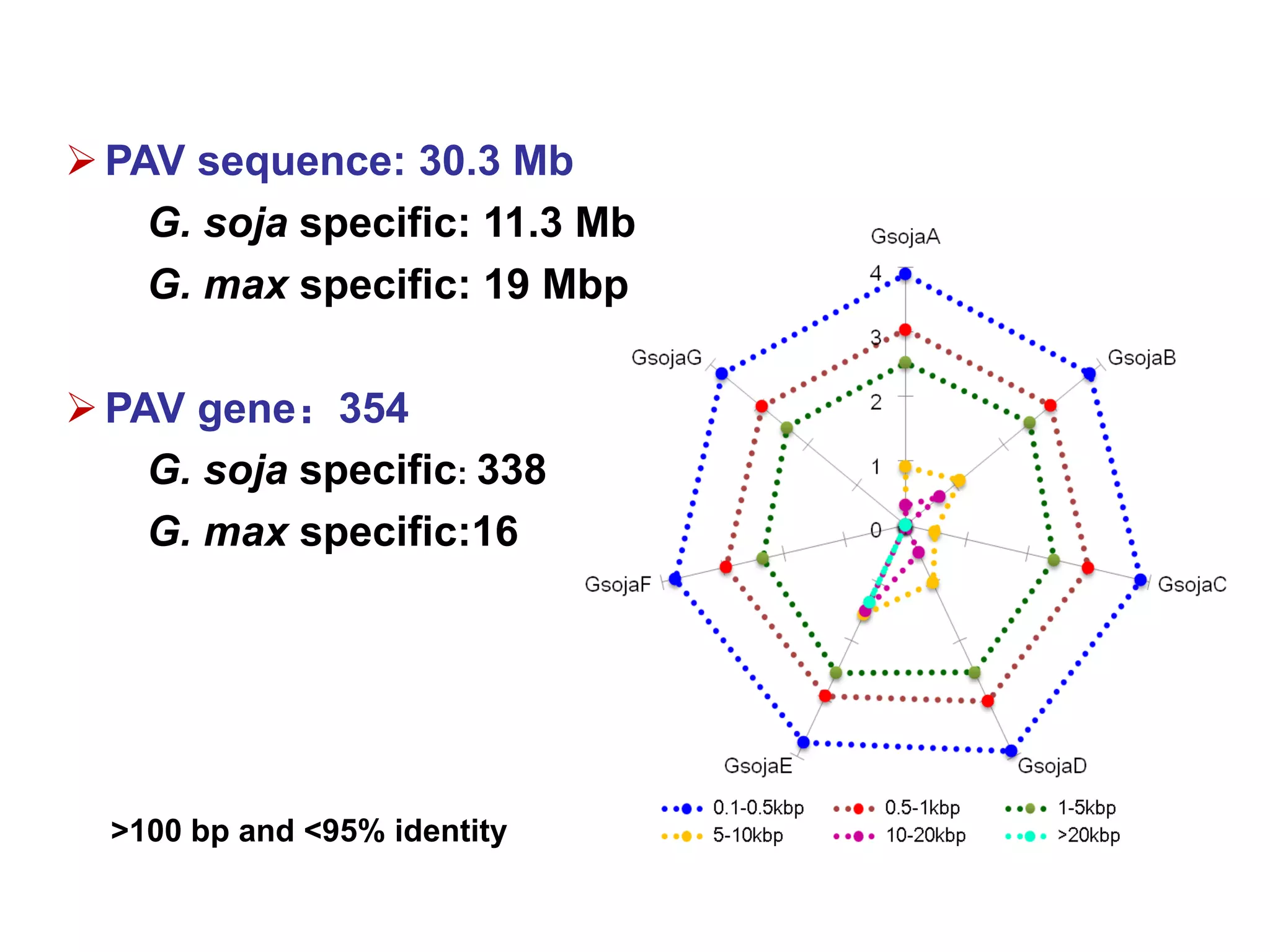
3) Comparison of G. soja and G. max genomes identified several million SNPs, indels, and presence/absence variants affecting genes related to domestication traits.