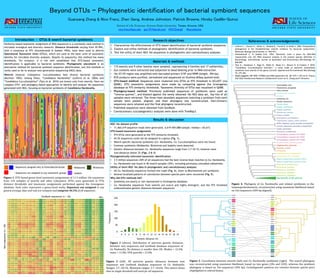
Zhang et al evol 2016 beyond otus phylogenetic identification of bacterial symbiont sequences
- 1. Beyond OTUs - Phylogenetic identification of bacterial symbiont sequences Guanyang Zhang & Nico Franz, Zhen Geng, Andrew Johnston, Patrick Browne, Hinsby Cadillo-Quiroz School of Life Sciences, Arizona State University, Tempe, Arizona, USA nico.franz@asu.edu gyz151@gmail.com @GYZhang2 @taxonbytes Similarity-based taxonomic assignment of DNA sequences is a commonly used method in microbial ecological and diversity research. Distance thresholds varying from 95-99%, with a consensus at 97% (Stackebrandt & Goebel 1994), have been used to delimit Operational Taxonomic Units (OTUs), which are used as the basic units of taxonomic identity for microbial diversity analyses. Despite its popularity, this method has many drawbacks. For instance, it is not well established that OTU-based taxonomic identification is applicable to bacterial symbionts. Phylogenetic placement is an alternative method for bacterial symbiont sequence identification, but this methods is rarely used so far to analyze next generation sequencing (NGS) data. Weevils (Insecta: Coleoptera: Curculionoidea) host diverse bacterial symbionts (Buchner 1965). Among these, “Candidatus Nardonella” (Lefèvre et al. 2004) and “Candidatus Curculioniphilus” (Toju et al. 2010) are known only from weevils. Here we compare OTU- and phylogeny-based approaches to identify and analyze 16s sequences generated with NGS, focusing on bacterial symbionts of Candidatus Nardonella. Introduction – OTUs & weevil bacterial symbionts 1. Characterize the effectiveness of OTU-based identification of bacterial symbiont sequences. 2. Explore and refine methods of phylogenetic identification of bacterial symbionts. 3. Test the utility of short NGS 16s sequences in phylogenetic and coevolutionary analyses. Research objectives • 115 weevils and 9 other beetles were sampled, representing 4 families and 17 subfamilies. • Gut contents were extracted and subjected to bead-beating prior to DNA extraction. • 16s V3-V5 region was amplified with barcoded primers 515F and 909R (length: 394 bp). • PCR products were purified, normalized and sequenced on Illumina MiSeq (paired-end). • OTU-based method: Sequences were clustered into OTUs at 97% threshold in UCLUST (via QIIME). OTU taxonomic assignments were made by comparing against the Greengenes database at 97% similarity thresholds. Taxonomic diversity of OTUs was visualized in QIIME. • Phylogeny-based method: Previously published sequences of symbionts were used as “reverse queries”, and blasted against the newly obtained 16s NGS data set. Top hits of all samples were retrieved. The three most abundant sequences matching a symbiont from each sample were pooled, aligned, and their phylogeny was reconstructed. Non-chimeric sequences were retained and the final phylogeny reconstructed. • Published sequences were obtained from GenBank. • Coevolutionary (cocladogenetic) analyses were done with TreeMap3. Materials & methods NGS 16s dataset profile • 11,464,419 sequence reads were generated. 4,619-459,088/sample, median = 65,615. OTU-based taxonomic assignments • 974 OTUs were generated at the 97% similarity threshold. • 44.5% sequences could not be assigned to a genus (Fig. 1). • Weevil-specific bacterial symbionts (Ca. Nardonella, Ca. Curculioniphilus) were not found. • Common symbionts (Wolbachia, Rickettsia and Sodalis) were detected. • Genetic distances between Ca. Nardonella sequences range from 3.7-18.1%; however none had distances below 3% (Figs. 2 & 3). Phylogenetically informed taxonomic identification • 3.5 million sequences (30% of all sequences) had the best reverse blast matches to Ca. Nardonella. • Ca. Nardonella was found in 40 weevil samples (35%), including previously unstudied subfamilies. Utility of short NGS 16s data in phylogenetic and coevolutionary analyses • All Ca. Nardonella sequences formed one clade (Fig. 4), sister to Blochmannia ant symbionts. • Several localized patterns of coevolution between species pairs were recovered (Fig. 5). Why did OTU methods fail? • Symbionts are poorly or not represented in Greengenes database. • Ca. Nardonella sequences from weevils are scarce and highly divergent; and the 97% threshold underestimates genetic distances between sequences. Results & discussion – Sequences assigned only to Enterobacteriaceae – Sequences not assigned to any taxonomic groups Figure 1. OTU-based genus-level taxonomic assignments of 11.5 million 16s sequences from 124 samples of weevils and other Coleoptera. OTUs were generated at 97% distance thresholds and taxonomic assignments performed against the Greengenes database. Each color represents a genus-level entity. Sequences not assigned to any genera (orange, blue and red) are rampant and comprise 44.5% of all sequences. Figure 3 (left). All pairwise genetic distances between new sequences and GenBank database sequences of Ca. Nadonella. Ranges: 3.7–18.1%. Minimum ranges: 3.7–10.6%. This matrix shows that no single threshold will work for all sequences. Figure 5. Coevolution between weevils (left) and Ca. Nardonella symbionts (right). The weevil phylogeny was reconstructed using maximum likelihood, based on two genes (28s and COII); whereas the symbiont phylogeny is based on 16s sequences (404 bp). Cocladogenetic patterns are common between species pairs (highlighted in colored boxes). Nardonella of 66 weevils Figure 4. Phylogeny of Ca. Nardonella and related symbionts in the Gammaproteobacteria, reconstructed using maximum likelihood based on 16s sequences (404 bp aligned). • Lefèvre C., Charles H., Vallier A., Delobel B., Farrell B. & Heddi A. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Molecular Biology and Evolution 21: 965–973. • Stackebrandt E. & Goebel B.M. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in Bacteriology. International Journal of Systematic and Evolutionary Microbiology 44: 846–849. • Toju H., Hosokawa T., Koga R., Nikoh N., Meng X.Y., Kimura N. & Fukatsu T. 2010. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Applied and Environmental Microbiology 76: 275–282. • Grant support: NSF DEB-1155984 and USDA Agreement No. 58-1275-1-335 (to N. Franz); ASU SOLS Postdoctoral Research Collaborative Grant (to G. Zhang and P. Browne). References & acknowledgements GenBank sequences (n = 26) Newsequences(n=40) Min. 11.7% 9.9% 10.2% 12.8% 11.3% 12.2% 12.5% 12.7% 11.5% 3.7% 10.2% 12.1% 13.0% 9.5% 12.0% 14.0% 9.3% 9.9% 5.0% 11.3% 12.0% 9.9% 5.0% 3.7% 9.3% 9.9% 3.7% 12.2% 11.0% 11.3% 13.3% 12.1% 13.0% 13.6% 13.8% 12.0% 3.9% 10.4% 12.9% 14.3% 10.8% 13.1% 14.8% 9.8% 10.4% 5.0% 12.1% 13.1% 11.0% 5.0% 4.2% 9.8% 10.4% 3.9% 11.4% 10.5% 10.7% 12.2% 10.8% 11.9% 12.5% 11.9% 11.0% 4.0% 9.7% 11.6% 12.5% 9.5% 11.7% 13.8% 9.8% 10.4% 5.3% 10.8% 11.7% 10.5% 5.3% 4.2% 9.8% 10.4% 4.0% 12.0% 10.8% 10.2% 13.3% 12.2% 13.5% 13.6% 13.0% 11.5% 4.0% 11.6% 13.0% 14.9% 11.9% 12.3% 14.9% 12.3% 9.4% 5.3% 12.2% 12.3% 10.8% 5.3% 4.5% 12.3% 9.4% 4.0% 12.2% 10.2% 10.5% 12.5% 11.9% 13.0% 12.5% 12.7% 10.7% 4.0% 9.9% 12.7% 13.3% 11.1% 12.0% 14.6% 9.6% 10.4% 4.5% 11.9% 12.0% 10.2% 4.5% 4.5% 9.6% 10.4% 4.0% 12.7% 11.0% 10.5% 14.1% 12.7% 13.5% 13.8% 12.4% 10.7% 4.2% 11.0% 12.9% 14.3% 11.4% 12.5% 14.8% 11.4% 9.6% 5.0% 12.7% 12.5% 11.0% 5.0% 4.5% 11.4% 9.6% 4.2% 11.1% 10.2% 10.5% 12.8% 11.1% 11.6% 13.0% 12.9% 11.0% 4.5% 9.7% 12.4% 13.3% 10.1% 12.0% 14.0% 9.0% 9.9% 4.7% 11.1% 12.0% 10.2% 4.7% 4.5% 9.0% 9.9% 4.5% 12.5% 10.8% 11.0% 13.3% 12.4% 12.7% 13.3% 13.5% 12.0% 5.0% 10.4% 12.9% 14.1% 10.6% 12.8% 14.6% 9.6% 10.4% 4.5% 12.4% 12.8% 10.8% 4.5% 5.3% 9.6% 10.4% 4.5% 11.7% 11.0% 11.3% 13.6% 11.3% 12.2% 13.6% 13.3% 11.8% 4.7% 10.5% 12.9% 13.8% 10.6% 12.5% 14.6% 9.6% 10.4% 5.0% 11.3% 12.5% 11.0% 5.0% 4.7% 9.6% 10.4% 4.7% 13.0% 11.8% 11.3% 14.1% 12.7% 13.5% 13.3% 15.7% 13.1% 5.2% 12.6% 14.0% 15.4% 13.0% 13.3% 16.9% 12.2% 12.0% 6.9% 12.7% 13.3% 11.8% 6.9% 5.0% 12.2% 12.0% 5.0% 12.8% 11.1% 11.6% 13.6% 12.4% 13.3% 13.1% 13.0% 12.1% 5.0% 11.3% 13.5% 14.1% 10.9% 13.1% 14.3% 9.9% 11.0% 6.1% 12.4% 13.1% 11.1% 6.1% 5.0% 9.9% 11.0% 5.0% 11.4% 10.8% 11.0% 13.6% 11.1% 12.7% 12.8% 14.4% 11.0% 5.5% 10.5% 13.7% 13.8% 12.4% 12.0% 15.1% 12.0% 12.0% 5.8% 11.1% 12.0% 10.8% 5.8% 5.0% 12.0% 12.0% 5.0% 13.0% 11.3% 11.0% 13.8% 12.9% 13.2% 13.3% 13.5% 11.5% 5.2% 10.5% 12.9% 14.6% 10.8% 12.8% 14.8% 9.8% 10.9% 6.1% 12.9% 12.8% 11.3% 6.1% 5.5% 9.8% 10.9% 5.2% 14.9% 12.9% 13.4% 16.5% 14.8% 15.6% 15.7% 15.5% 11.5% 5.3% 11.3% 16.2% 16.4% 14.0% 15.7% 15.6% 13.0% 12.0% 5.5% 14.8% 15.7% 12.9% 5.5% 5.3% 13.0% 12.0% 5.3% 14.3% 11.8% 12.1% 14.6% 14.0% 15.6% 13.8% 14.6% 12.8% 5.5% 12.8% 15.1% 14.1% 13.5% 15.2% 17.2% 11.2% 13.3% 6.3% 14.0% 15.2% 11.8% 6.3% 5.3% 11.2% 13.3% 5.3% 13.8% 11.9% 11.8% 14.2% 13.5% 14.6% 13.6% 13.1% 11.3% 5.6% 11.6% 14.1% 14.1% 12.2% 13.9% 15.7% 11.8% 10.7% 7.2% 13.5% 13.9% 11.9% 7.2% 5.3% 11.8% 10.7% 5.3% 11.9% 10.5% 10.5% 13.0% 11.6% 11.9% 12.8% 12.2% 12.8% 5.8% 11.5% 12.4% 12.7% 9.8% 12.3% 14.0% 9.8% 11.7% 5.8% 11.6% 12.3% 10.5% 5.8% 5.8% 9.8% 11.7% 5.8% 13.3% 11.6% 10.7% 14.9% 13.2% 13.2% 13.8% 12.7% 10.4% 6.3% 10.7% 13.2% 14.3% 13.0% 14.1% 15.9% 10.6% 12.0% 6.3% 13.2% 14.1% 11.6% 6.3% 6.1% 10.6% 12.0% 6.1% 11.9% 10.2% 10.2% 12.8% 11.3% 12.2% 12.5% 12.2% 12.8% 6.3% 12.1% 12.1% 13.0% 10.3% 12.3% 13.5% 11.2% 12.8% 6.1% 11.3% 12.3% 10.2% 6.1% 6.6% 11.2% 12.8% 6.1% 10.9% 9.4% 8.9% 12.5% 10.8% 11.9% 12.3% 12.5% 12.9% 6.3% 12.4% 11.6% 13.6% 11.1% 11.2% 14.9% 11.5% 10.7% 8.2% 10.8% 11.2% 9.4% 8.2% 6.6% 11.5% 10.7% 6.3% 9.1% 10.8% 9.5% 10.7% 9.0% 10.4% 10.5% 6.4% 13.1% 14.8% 11.7% 7.8% 12.3% 6.6% 9.9% 14.0% 13.8% 13.0% 15.1% 9.0% 9.9% 10.8% 15.1% 14.5% 13.8% 13.0% 6.4% 13.2% 13.0% 12.7% 14.0% 13.4% 12.9% 13.5% 14.3% 8.0% 13.6% 6.4% 12.7% 14.2% 13.1% 13.7% 14.5% 8.8% 7.8% 13.1% 13.4% 13.7% 13.0% 13.1% 14.2% 8.8% 7.8% 6.4% 12.0% 11.6% 11.0% 14.7% 12.4% 13.3% 13.3% 14.4% 14.7% 6.6% 14.0% 14.1% 15.2% 11.7% 12.6% 15.6% 12.0% 12.6% 9.0% 12.4% 12.6% 11.6% 9.0% 7.1% 12.0% 12.6% 6.6% 14.6% 13.7% 13.1% 14.6% 14.0% 14.8% 14.4% 14.6% 10.4% 6.8% 11.3% 12.7% 15.4% 12.4% 14.7% 14.8% 12.8% 12.0% 7.7% 14.0% 14.7% 13.7% 7.7% 7.6% 12.8% 12.0% 6.8% 14.6% 13.7% 13.1% 14.6% 14.0% 14.8% 14.4% 14.6% 10.4% 6.8% 11.3% 12.7% 15.4% 12.4% 14.7% 14.8% 12.8% 12.0% 7.7% 14.0% 14.7% 13.7% 7.7% 7.6% 12.8% 12.0% 6.8% 13.0% 11.3% 11.5% 14.4% 12.9% 14.3% 13.6% 14.1% 13.6% 6.9% 13.9% 13.7% 14.6% 11.9% 12.8% 14.8% 11.7% 11.7% 9.2% 12.9% 12.8% 11.3% 9.2% 6.9% 11.7% 11.7% 6.9% 13.1% 12.4% 12.4% 13.7% 13.3% 12.8% 14.0% 13.1% 7.8% 13.6% 7.0% 12.1% 14.2% 12.6% 13.5% 13.9% 8.5% 7.5% 12.8% 13.3% 13.5% 12.4% 12.8% 13.6% 8.5% 7.5% 7.0% 13.1% 13.0% 12.4% 15.2% 13.5% 14.4% 13.9% 14.4% 14.5% 7.1% 14.8% 14.6% 15.2% 13.0% 13.7% 15.7% 13.1% 12.9% 9.0% 13.5% 13.7% 13.0% 9.0% 7.1% 13.1% 12.9% 7.1% 13.9% 13.5% 12.9% 16.0% 14.6% 15.4% 15.5% 15.8% 15.3% 7.4% 15.4% 15.4% 17.1% 13.3% 12.1% 16.8% 13.9% 12.9% 9.5% 14.6% 12.1% 13.5% 9.5% 7.1% 13.9% 12.9% 7.1% 13.5% 12.4% 11.8% 15.7% 13.5% 14.6% 16.0% 13.3% 13.9% 7.6% 13.1% 14.0% 17.0% 12.7% 14.1% 15.6% 12.5% 11.7% 8.4% 13.5% 14.1% 12.4% 8.4% 7.9% 12.5% 11.7% 7.6% 13.0% 13.7% 13.2% 16.3% 13.5% 13.0% 16.3% 16.3% 15.8% 7.9% 15.1% 14.9% 16.2% 12.7% 13.9% 17.2% 13.3% 13.9% 8.2% 13.5% 13.9% 13.7% 8.2% 7.9% 13.3% 13.9% 7.9% 9.9% 9.5% 8.4% 11.6% 9.9% 10.5% 10.8% 11.5% 8.3% 11.2% 8.8% 8.9% 11.3% 10.5% 10.5% 11.3% 8.3% 8.3% 12.6% 9.9% 10.5% 9.5% 12.6% 11.0% 8.3% 8.3% 8.3% 9.6% 11.9% 10.0% 10.4% 9.8% 10.9% 10.4% 8.9% 14.4% 15.8% 13.6% 8.6% 12.3% 8.8% 9.7% 14.5% 13.2% 14.1% 17.7% 9.8% 9.7% 11.9% 17.7% 15.5% 13.2% 14.1% 8.6% 12.0% 12.7% 12.1% 15.2% 12.5% 13.3% 14.7% 13.6% 13.7% 8.7% 13.7% 13.6% 14.9% 11.7% 12.6% 15.4% 12.8% 12.1% 11.1% 12.5% 12.6% 12.7% 11.1% 8.7% 12.8% 12.1% 8.7% 10.4% 9.7% 9.4% 14.1% 10.1% 11.7% 13.3% 13.6% 14.7% 8.7% 12.9% 12.7% 14.1% 10.1% 11.5% 15.9% 11.5% 13.1% 9.8% 10.1% 11.5% 9.7% 9.8% 8.7% 11.5% 13.1% 8.7% 12.8% 13.0% 12.4% 15.5% 13.3% 13.6% 14.4% 13.3% 13.4% 9.0% 14.0% 13.8% 15.5% 12.0% 12.9% 14.6% 13.1% 12.6% 10.9% 13.3% 12.9% 13.0% 10.9% 9.0% 13.1% 12.6% 9.0% 13.9% 13.2% 12.7% 15.9% 13.9% 14.7% 16.4% 15.6% 15.0% 14.9% 14.2% 13.2% 17.4% 16.0% 14.6% 15.5% 10.6% 9.9% 16.3% 13.9% 14.6% 13.2% 16.3% 14.7% 10.6% 9.9% 9.9% 11.0% 10.6% 10.1% 15.0% 11.2% 12.6% 13.9% 14.8% 13.4% 15.3% 13.9% 12.7% 16.3% 13.9% 11.1% 14.6% 13.0% 10.7% 15.3% 11.2% 11.1% 10.6% 15.3% 14.8% 13.0% 10.7% 10.1% 10.3% 11.0% 10.2% 13.8% 10.5% 11.3% 13.5% 13.5% 13.4% 14.0% 13.7% 11.3% 14.6% 11.9% 10.9% 15.4% 12.8% 12.3% 14.3% 10.5% 10.9% 11.0% 14.3% 13.8% 12.8% 12.3% 10.2% 14.1% 14.2% 13.1% 17.1% 14.6% 14.1% 17.0% 15.8% 13.9% 10.6% 15.0% 15.4% 18.1% 13.5% 14.4% 15.4% 13.6% 14.4% 11.4% 14.6% 14.4% 14.2% 11.4% 10.6% 13.6% 14.4% 10.6% Figure 2 (above). Distribution of pairwise genetic distances between new sequences and GenBank database sequences of Ca. Nadonella. No distance is smaller than 3%. Median = 12.4%; mean = 11.8%; 95% quartile = 15.4%. Genetic distance (%) Frequency – Wolbachia – Sodalis – Rickettsia
