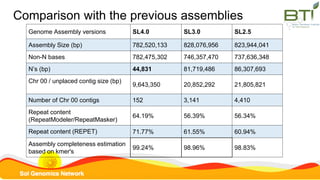
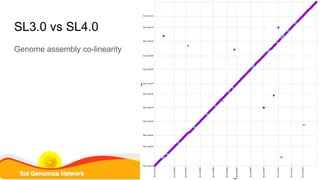
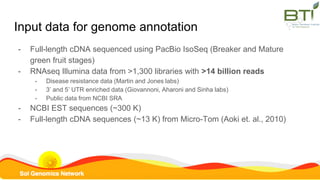
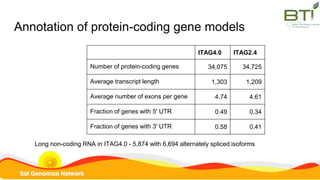
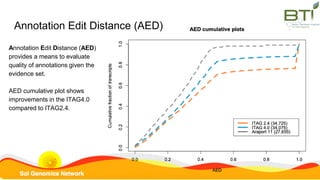
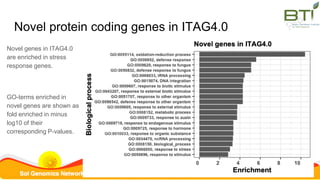
This document summarizes the Solanum lycopersicum Heinz 1706 genome assembly SL4.0 and its annotation ITAG4.0 from the Sol Genomics Network. Key points include: SL4.0 has an improved assembly compared to previous versions with longer contigs and less unplaced sequence. It was generated using PacBio and Illumina sequencing. The annotation ITAG4.0 contains 34,075 protein-coding genes compared to 34,725 in the previous version, with improvements in 5' and 3' UTR identification. Novel genes in ITAG4.0 are enriched for stress response functions. Validation was performed using various omics datasets and optical mapping.